4RLT
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Fisetin | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 3,7,3',4'-TETRAHYDROXYFLAVONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
7EZ2
 
 | | Holo L-16 ScaI Tetrahymena ribozyme | | Descriptor: | Holo L-16 ScaI Tetrahymena ribozyme, Holo L-16 ScaI Tetrahymena ribozyme S1, Holo L-16 ScaI Tetrahymena ribozyme S2, ... | | Authors: | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
8FLQ
 
 | | Human PTH1R in complex with PTH(1-34) and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5KTO
 
 | | Crystal structure of Pyrococcus horikoshii quinolinate synthase (NadA) with bound quinolinate and Fe4S4 cluster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fenwick, M.K, Ealick, S.E. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Crystal Structures of the Iron-Sulfur Cluster-Dependent Quinolinate Synthase in Complex with Dihydroxyacetone Phosphate, Iminoaspartate Analogues, and Quinolinate.
Biochemistry, 55, 2016
|
|
6BBX
 
 | | Crystal structure of TnmS3 in complex with TNM C | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2R,3R)-2,3-dihydroxy-3-[(1aS,11S,11aR,14Z,18R)-3,7,8,18-tetrahydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]butanoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-19 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
6X9F
 
 | | Pseudomonas aeruginosa MurC with AZ8074 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ8074
To Be Published
|
|
8CVR
 
 | | Human 20S proteasome with MG-132 | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-4-methyl-1-oxopentan-2-yl]-L-leucinamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6XB2
 
 | | Room temperature X-ray crystallography reveals catalytic cysteine in the SARS-CoV-2 3CL Mpro is highly reactive: Insights for enzyme mechanism and drug design | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kneller, D.W, Kovalevsky, A, Coates, L. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Room-temperature X-ray crystallography reveals the oxidation and reactivity of cysteine residues in SARS-CoV-2 3CL M pro : insights into enzyme mechanism and drug design.
Iucrj, 7, 2020
|
|
8IR1
 
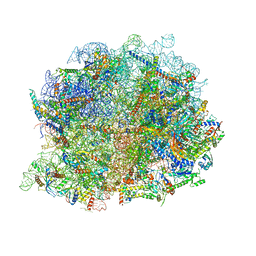 | | human nuclear pre-60S ribosomal particle - State A | | Descriptor: | 28S rRNA, 5.8S rRMA, 5S RNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-17 | | Release date: | 2023-08-09 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
8XQT
 
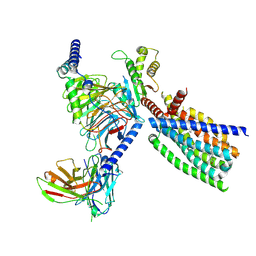 | | Structure of human class T GPCR TAS2R14-Gi complex. | | Descriptor: | CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Pei, Y, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
6XH0
 
 | |
9IYH
 
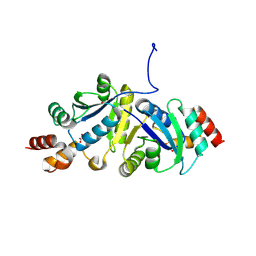 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution.
To Be Published
|
|
8FBM
 
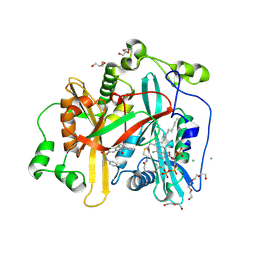 | |
6XMX
 
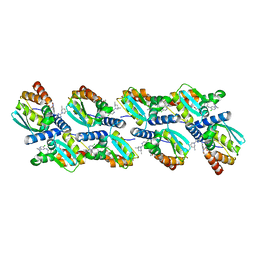 | | Cryo-EM structure of BCL6 bound to BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Yoon, H, Burman, S.S.R, Hunkeler, M, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2020-07-01 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Small-molecule-induced polymerization triggers degradation of BCL6.
Nature, 588, 2020
|
|
9IYF
 
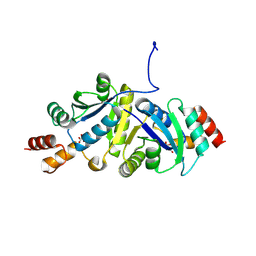 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.37 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.37 A resolution.
To Be Published
|
|
5KVE
 
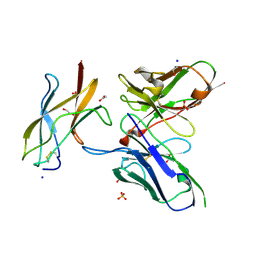 | | Zika specific antibody, ZV-48, bound to ZIKA envelope DIII | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Genome polyprotein, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
8FOW
 
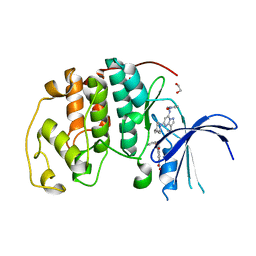 | | Ternary complex of CDK2 with small molecule ligands TW8672 and Dinaciclib | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-01-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
9IYE
 
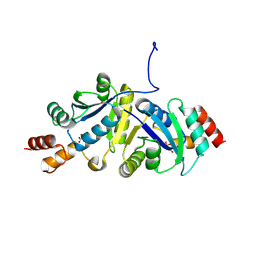 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution.
To Be Published
|
|
9LU9
 
 | | The chimeric flagellar motor complex between MotA1B1 from Paenibacillus sp. TCA20 and MotAB from E.coli, state 1 | | Descriptor: | Flagellar motor protein MotA, Lauryl Maltose Neopentyl Glycol, MotB1,Motility protein B | | Authors: | Onoe, S, Nishikino, T, Kinoshita, M, Kishikawa, J, Kato, T. | | Deposit date: | 2025-02-08 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Structure of the Flagellar Motor Complex from Paenibacillus sp. TCA20.
Biomolecules, 15, 2025
|
|
8ZFT
 
 | |
6NFO
 
 | |
8QUG
 
 | | KRAS-G12C in Complex with Compound 1 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[2-[(2S)-2-methyl-1,4-diazepan-1-yl]pyrimidin-4-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fischer, G, Kratochvil, B. | | Deposit date: | 2023-10-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8C7W
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2304 | | Descriptor: | 1,2-ETHANEDIOL, 6-methyl-9-piperazin-1-yl-4-(3,4,5-trimethoxyphenyl)-5,7-dihydropyrido[4,3-d][2]benzazepine, Activin receptor type I, ... | | Authors: | Cros, J, Williams, E.P, Sweeney, M.N, Smil, D, Gonzalez-Alvarez, H, Al-awar, R, Bullock, A.N. | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Conformationally Constrained ALK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9KNE
 
 | | Crystal structure of human ERRg LBD in complex with 5-nitroindole | | Descriptor: | 5-nitro-1H-indole, Estrogen-related receptor gamma, Nuclear receptor-interacting protein 1, ... | | Authors: | Xu, T, Wang, N, Liu, J. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of indoles as potential endogenous ligands of ERR gamma and their modulation on drug binding.
Acta Pharmacol.Sin., 46, 2025
|
|
