5M33
 
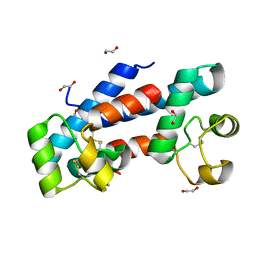 | | Structural tuning of CD81LEL (space group P21) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
6X7D
 
 | |
7UBJ
 
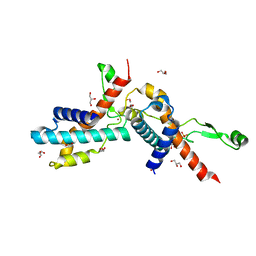 | | Transcription antitermination factor Qlambda, type-I crystal | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, CHLORIDE ION, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9HVJ
 
 | |
7UBL
 
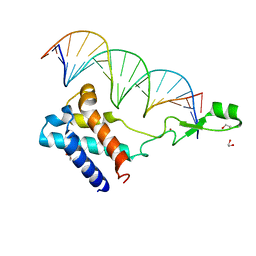 | | Transcription antitermination factor Qlambda in complex with Q-lambda-binding-element DNA | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, DNA (5'-D(P*CP*AP*CP*CP*CP*AP*AP*TP*TP*TP*TP*AP*TP*TP*CP*AP*AP*TP*G)-3'), ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8A5J
 
 | | Crystal structure of Human STE20-like kinase 1, MST1 in complex with compound XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, Serine/threonine-protein kinase 4 37kDa subunit | | Authors: | Nawrotek, A, Vuillard, L, Miallau, L. | | Deposit date: | 2022-06-15 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Crystal structure of the Kelch domain of human Keap1in complex with ligand S217879
To Be Published
|
|
6X7P
 
 | | LnmK in complex with 2-sulfonate-propionyl-oxa(dethia)-CoA | | Descriptor: | (2~{R})-1-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethoxy]-1-oxidanylidene-propane-2-sulfonic acid, (2~{S})-1-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethoxy]-1-oxidanylidene-propane-2-sulfonic acid, Conserved hypthetical protein, ... | | Authors: | Stunkard, L.M, Kick, B.J, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of LnmK, a Bifunctional Acyltransferase/Decarboxylase, with Substrate Analogues Reveal the Basis for Selectivity and Stereospecificity.
Biochemistry, 60, 2021
|
|
8RZC
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-imidazol-1-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
6HRZ
 
 | | EthR2 in complex with compound 4 (BDM72170) | | Descriptor: | 1-(1,3-benzothiazol-2-yl)guanidine, Probable transcriptional regulatory protein | | Authors: | Wintjens, R, Wohlkonig, A, Tanina, A. | | Deposit date: | 2018-09-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A fragment-based approach towards the discovery of N-substituted tropinones as inhibitors of Mycobacterium tuberculosis transcriptional regulator EthR2.
Eur J Med Chem, 167, 2019
|
|
6R2A
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD cocrystallized with succinylphosphonate phosphonoethyl ester (PESP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanyl-butanoic acid, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6HSE
 
 | | Structure of dithionite-reduced RsrR in spacegroup P2(1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Rrf2 family transcriptional regulator, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Transcription Regulator RsrR Reveals a [2Fe-2S] Cluster Coordinated by Cys, Glu, and His Residues.
J. Am. Chem. Soc., 141, 2019
|
|
6U0K
 
 | | TTBK2 kinase domain in complex with Compound 1 | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Marcotte, D.J, Chodaparambil, J.V. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | The crystal structure of the catalytic domain of tau tubulin kinase 2 in complex with a small-molecule inhibitor
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8RV7
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 4 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-oxidanylprop-1-ynyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
6R4C
 
 | | Aurora-A in complex with shape-diverse fragment 57 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, CHLORIDE ION, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
6X6H
 
 | |
7BE7
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
9HVM
 
 | | In-cell Structure of Pyrenoid Rubisco | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small subunit, chloroplastic 1 | | Authors: | Nadav, E, Zhen, H, Maud, D, Alireza, R, Juan R, P, Peijun, P. | | Deposit date: | 2024-12-30 | | Release date: | 2025-03-05 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | In-cell structure and variability of pyrenoid Rubisco.
Nat Commun, 16, 2025
|
|
7YGG
 
 | |
7BL1
 
 | | human complex II-BATS bound to membrane-attached Rab5a-GTP | | Descriptor: | Beclin-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tremel, S, Morado, D.R, Kovtun, O, Williams, R.L, Briggs, J.A.G, Munro, S, Ohashi, Y, Bertram, J, Perisic, O. | | Deposit date: | 2021-01-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes.
Nat Commun, 12, 2021
|
|
8YHW
 
 | | The Crystal Structure of NF-kB-inducing Kinase (NIK) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, MAGNESIUM ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of NF-kB-inducing Kinase (NIK) from Biortus
To Be Published
|
|
8PQZ
 
 | | Cytoplasmic dynein-1 A1/A2 motor domains bound to LIS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
7BGP
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in absence of DTT. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7UO0
 
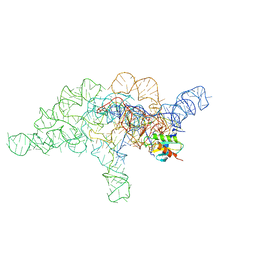 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, Precursor tRNA substrate G(-1) G(-2), RNase P RNA, ... | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
6XH3
 
 | |
6REE
 
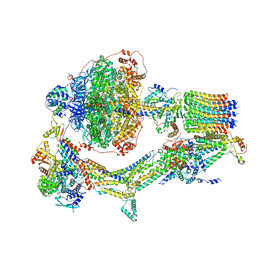 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3B, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
