8RCG
 
 | | W-formate dehydrogenase M405S from Desulfovibrio vulgaris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural and biochemical characterization of the M405S variant of Desulfovibrio vulgaris formate dehydrogenase.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
6HA7
 
 | | Crystal structure of the BiP NBD and MANF complex | | Descriptor: | 1,2-ETHANEDIOL, Endoplasmic reticulum chaperone BiP, Mesencephalic astrocyte-derived neurotrophic factor | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP.
Nat Commun, 10, 2019
|
|
6ZHW
 
 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 1 ps structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Branden, G, Neutze, R. | | Deposit date: | 2020-06-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
7TEM
 
 | | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis
To Be Published
|
|
7SQ7
 
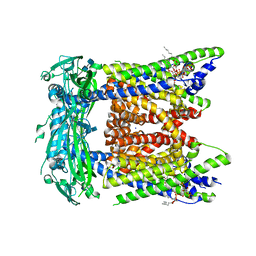 | | Cryo-EM structure of mouse PI(3,5)P2-bound TRPML1 channel at 2.41 Angstrom resolution | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZLU
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-2-deoxy-2-fluoro-D-glucose | | Descriptor: | 1,3-PROPANDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Ibba, R. | | Deposit date: | 2022-04-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
5YRB
 
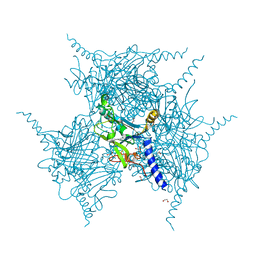 | | Crystal Structure of Oxidized Cypovirus Polyhedra R13A/E73C/Y83C/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
4RJ5
 
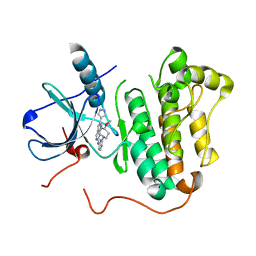 | | EGFR kinase (T790M/L858R) with inhibitor compound 5 | | Descriptor: | Epidermal growth factor receptor, N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-2-(1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridin-6-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
5NJC
 
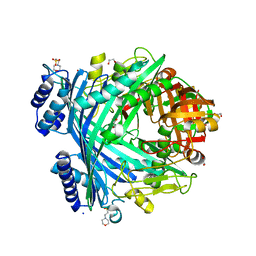 | | E. coli Microcin-processing metalloprotease TldD/E (TldD E263A mutant) with hexapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metalloprotease PmbA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
8F4U
 
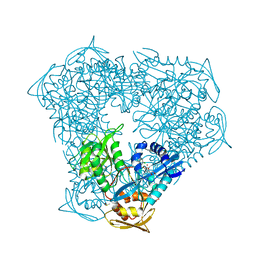 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with azelastine | | Descriptor: | 4-[(4-chlorophenyl)methyl]-2-[(4S)-1-methylazepan-4-yl]phthalazin-1(2H)-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
6NV7
 
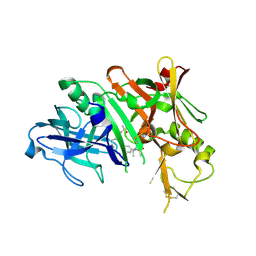 | | BACE1 in complex with a macrocyclic inhibitor | | Descriptor: | (E)-N-(2-methylpropylidene)-N~2~-{[(4S)-17-[(methylsulfonyl)(propyl)amino]-2-oxo-3-azatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaen-4-yl]methyl}-D-threoninamide, Beta-secretase 1 | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-02-04 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.132 Å) | | Cite: | Development of an Efficient Enzyme Production and Structure-Based Discovery Platform for BACE1 Inhibitors.
Biochemistry, 58, 2019
|
|
5NZ3
 
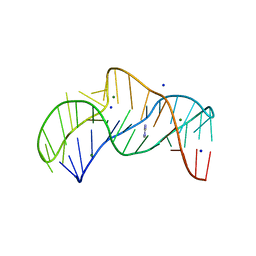 | | The structure of the thermobifida fusca guanidine III riboswitch with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
7YMU
 
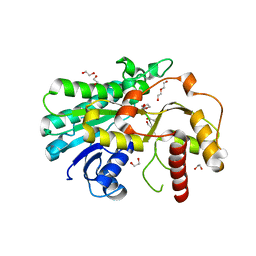 | | Structure of Alcohol dehydrogenase from [Candida] glabrata | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sun, Z.W, Liu, Y.F, Xu, G.C, Ni, Y. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rationla design of CgADH from candida glarata for asymmetric reduction of azacycolne.
To Be Published
|
|
8OEG
 
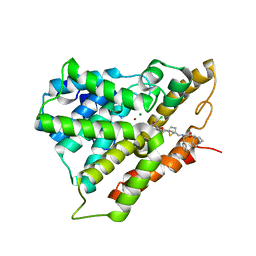 | | PDE4B bound to MAPI compound 92a | | Descriptor: | MAGNESIUM ION, ZINC ION, [(1~{S})-2-[3,5-bis(chloranyl)-1-oxidanyl-pyridin-4-yl]-1-(3,4-dimethoxyphenyl)ethyl] 5-[[[(1~{R})-2-[[(3~{R})-1-azabicyclo[2.2.2]octan-3-yl]oxy]-2-oxidanylidene-1-phenyl-ethyl]amino]methyl]thiophene-2-carboxylate, ... | | Authors: | Rizzi, A, Armani, E. | | Deposit date: | 2023-03-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of M 3 Antagonist-PDE4 Inhibitor (MAPI) Dual Pharmacology Molecules for the Treatment of COPD.
J.Med.Chem., 66, 2023
|
|
7C2V
 
 | | Crystal Structure of IRAK4 kinase in complex with the inhibitor CA-4948 | | Descriptor: | 2-(2-methylpyridin-4-yl)-N-[2-morpholin-4-yl-5-[(3R)-3-oxidanylpyrrolidin-1-yl]-[1,3]oxazolo[4,5-b]pyridin-6-yl]-1,3-oxazole-4-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Krishnamurthy, N.R, Robert, B. | | Deposit date: | 2020-05-09 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of CA-4948, an Orally Bioavailable IRAK4 Inhibitor for Treatment of Hematologic Malignancies.
Acs Med.Chem.Lett., 11, 2020
|
|
8EHP
 
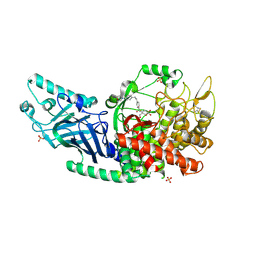 | | Co-crystal structure of Chaetomium glucosidase with compound 13 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(4-{[4-(morpholin-4-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6XUF
 
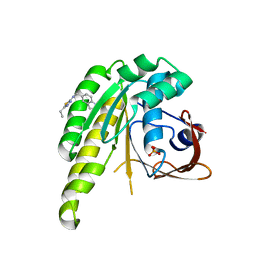 | | HumRadA1 in complex with 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine in P21 | | Descriptor: | 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Scott, D.E, Hyvonen, M.E. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.241 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To be published
|
|
6GL2
 
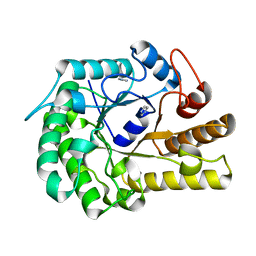 | | Structure of ZgEngAGH5_4 wild type at 1.2 Angstrom resolution | | Descriptor: | Endoglucanase, family GH5, IMIDAZOLE | | Authors: | Dorival, J, Ruppert, S, Gunnoo, M, Orlowski, A, Chapelais, M, Dabin, J, Labourel, A, Thompson, D, Michel, G, Czjzek, M, Genicot, S. | | Deposit date: | 2018-05-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The laterally acquired GH5ZgEngAGH5_4from the marine bacteriumZobellia galactanivoransis dedicated to hemicellulose hydrolysis.
Biochem. J., 475, 2018
|
|
5YRA
 
 | | Crystal Structure of Cypovirus Polyhedra R13A/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
6GOZ
 
 | | Structure of mEos4b in the green long-lived dark state | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | Deposit date: | 2018-06-04 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Mechanistic Investigations of Green mEos4b Reveal a Dynamic Long-Lived Dark State.
J.Am.Chem.Soc., 2020
|
|
6BED
 
 | | Crystal structure of VACV D13 in complex with Rifampicin | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, RIFAMPICIN, ... | | Authors: | Garriga, D, Accurso, C, Coulibaly, F. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the inhibition of poxvirus assembly by the antibiotic rifampicin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8PBU
 
 | | Mutant K1482M of the dihydroorotase domain of human CAD protein bound to the inhibitor fluoorotate | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD protein, FORMIC ACID, ... | | Authors: | del Cano-Ochoa, F, Ramon-Maiques, S. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Beyond genetics: Deciphering the impact of missense variants in CAD deficiency.
J Inherit Metab Dis, 46, 2023
|
|
8EPR
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 19 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[3-({[(5M)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EUT
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 27 | | Descriptor: | (2R,3R,4R,5S)-1-[8-(furan-2-yl)octyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8JPC
 
 | | cryo-EM structure of NTSR1-GRK2-Galpha(q) complexes 2 | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-adrenergic receptor kinase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
