5HVJ
 
 | |
5M5U
 
 | |
9KNT
 
 | | ERDRP-0519-bound measles virus L-P complex | | Descriptor: | 2-methyl-~{N}-[4-[(2~{S})-2-(2-morpholin-4-ylethyl)piperidin-1-yl]sulfonylphenyl]-5-(trifluoromethyl)pyrazole-3-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Wang, Y.R, Zhang, H.Q. | | Deposit date: | 2024-11-19 | | Release date: | 2025-07-16 | | Last modified: | 2025-09-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the measles virus polymerase complex with non-nucleoside inhibitors and mechanism of inhibition.
Cell, 188, 2025
|
|
6Q4M
 
 | | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, PHOSPHATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Gundogdu, M, van Aalten, D.M.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation
To Be Published
|
|
6QDF
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000096 | | Descriptor: | 3-[[6-tert-butyl-2-[methyl-[(3S)-1-methylpyrrolidin-3-yl]amino]thieno[3,2-d]pyrimidin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
8PIA
 
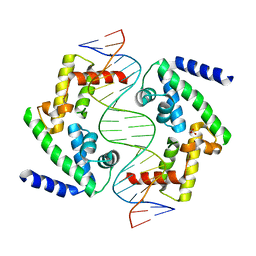 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -181G>T) | | Descriptor: | Chains: E, Chains: F, GLYCEROL, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
5HK4
 
 | | Structure function studies of R. palustris RubisCO (A47V-M331A mutant) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
9KNZ
 
 | | ERDRP-0519-bound Nipah virus L-P complex | | Descriptor: | 2-methyl-~{N}-[4-[(2~{S})-2-(2-morpholin-4-ylethyl)piperidin-1-yl]sulfonylphenyl]-5-(trifluoromethyl)pyrazole-3-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Wang, Y.R, Zhang, H.Q. | | Deposit date: | 2024-11-19 | | Release date: | 2025-07-16 | | Last modified: | 2025-09-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the measles virus polymerase complex with non-nucleoside inhibitors and mechanism of inhibition.
Cell, 188, 2025
|
|
9KNV
 
 | | AS-136A-bound measles virus L-P complex | | Descriptor: | 2-methyl-~{N}-(4-piperidin-1-ylsulfonylphenyl)-5-(trifluoromethyl)pyrazole-3-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Wang, Y.R, Zhang, H.Q. | | Deposit date: | 2024-11-19 | | Release date: | 2025-07-16 | | Last modified: | 2025-09-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the measles virus polymerase complex with non-nucleoside inhibitors and mechanism of inhibition.
Cell, 188, 2025
|
|
5JWH
 
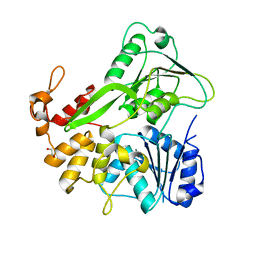 | | Apo structure | | Descriptor: | 1,2-ETHANEDIOL, NS3 helicase | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5M0I
 
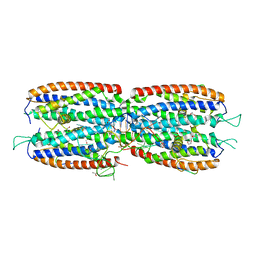 | | Crystal structure of the nuclear complex with She2p and the ASH1 mRNA E3-localization element | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ASH1-E3 element, ... | | Authors: | Edelmann, F.T, Janowski, R, Niessing, D. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8PI8
 
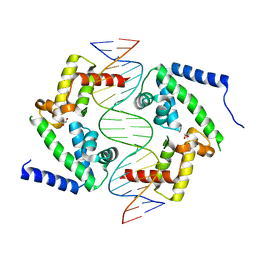 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA | | Descriptor: | Chains: E, Chains: F, GLYCEROL, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
8BAD
 
 | | Tpp80Aa1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Binary toxin A-like protein, CALCIUM ION, ... | | Authors: | Best, H.L, Williamson, L.J, Rizkallah, P.J, Berry, C, Lloyd-Evans, E. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure of Bacillus thuringiensis Tpp80Aa1 and Its Interaction with Galactose-Containing Glycolipids.
Toxins, 14, 2022
|
|
4R3Z
 
 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8PJG
 
 | | F11 TCR in complex with Human Leukocyte Antigen class II allotype DR1 presenting P11T->R modified influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
8ZQW
 
 | | The crystal structure of PDE4D with isoaurostatin derivatives 2-9 | | Descriptor: | (3~{Z})-3-[[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylidene]-6-oxidanyl-1~{H}-indol-2-one, 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-06-03 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.100019 Å) | | Cite: | Structure-based optimization of isoaurostatin as novel PDE4 inhibitors with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
8ZQU
 
 | | The crystal structure of PDE4D with isoaurostatin derivatives 2-6 | | Descriptor: | (3~{Z})-3-[[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylidene]-6-oxidanyl-1-benzofuran-2-one, 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-06-03 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.00004554 Å) | | Cite: | Structure-based optimization of isoaurostatin as novel PDE4 inhibitors with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
4ZED
 
 | |
6XTW
 
 | | HumRadA33F in complex with peptidic inhibitor 6 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
8BE0
 
 | |
8E3S
 
 | |
8GDA
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (2S,5R)-5-[(1E,3S)-4,4-difluoro-3-hydroxy-4-phenylbut-1-en-1-yl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Jiangqian, M, Sheng, C, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BDR
 
 | |
6GIN
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with an Quinazolinone based ALK2 inhibitor with a 4-morpholinophenyl solvent accessible group. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-morpholin-4-ylphenyl)-6-quinolin-4-yl-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Kopec, J, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
4ZJP
 
 | | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose | | Descriptor: | 1,2-ETHANEDIOL, Monosaccharide-transporting ATPase, beta-D-ribopyranose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Glenn, A.S, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose
To be published
|
|
