6QBC
 
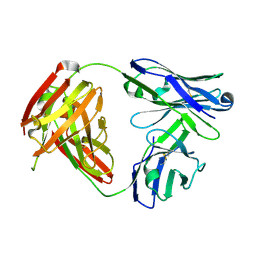 | | structure of anti-Mcl1 Fab | | Descriptor: | Anti-Mcl1 Fab Heavy Chain, Anti-Mcl1 Fab Light Chain | | Authors: | Luptak, J. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8CIQ
 
 | | JzTx-34 toxin peptide | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJP
 
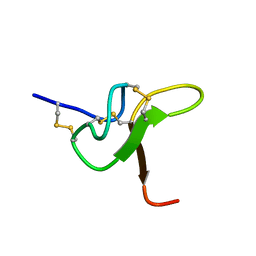 | | JzTx-34 toxin peptide H18A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
6IPG
 
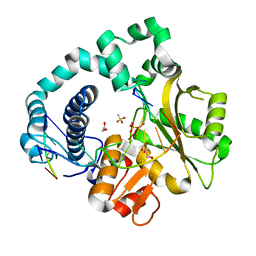 | | Post-catalytic Complex of Human DNA Polymerase Mu with Templating Cytosine and Mg-8oxodGMP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*CP*GP*GP*CP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-11-03 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
8CJS
 
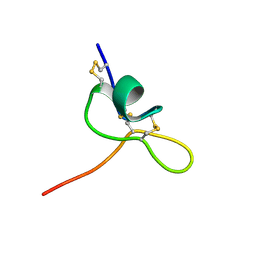 | | JzTx-34 toxin peptide W31A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
5LQV
 
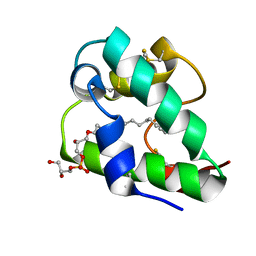 | | Spatial structure of the lentil lipid transfer protein in complex with anionic lysolipid LPPG | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], Non-specific lipid-transfer protein 2 | | Authors: | Mineev, K.S, Shenkarev, Z.O, Arseniev, A.S, Melnikova, D.N, Finkina, E.I, Ovchinnikova, T.V. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Ligand Binding Properties of the Lentil Lipid Transfer Protein: Molecular Insight into the Possible Mechanism of Lipid Uptake.
Biochemistry, 56, 2017
|
|
8CJQ
 
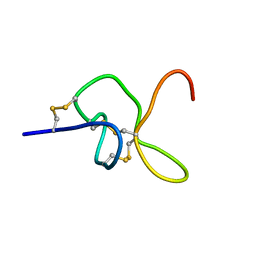 | | JzTx-34 toxin peptide E20A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJR
 
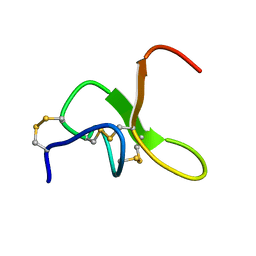 | | JzTx-34 toxin peptide W25A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8F5X
 
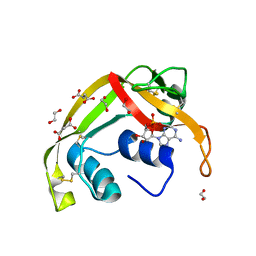 | | Crystal structure of human eosinophil-derived neurotoxin (EDN, ribonuclease 2) in complex with 5'-adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Non-secretory ribonuclease, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
8CJT
 
 | | JzTx-34 toxin peptide W33A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
9I33
 
 | |
6E6T
 
 | | Dieckmann cyclase, NcmC, bound to cerulenin | | Descriptor: | (4S,5R)-4,5-dihydroxy-5-[(3E,6E)-octa-3,6-dien-1-yl]pyrrolidin-2-one, NcmC, SULFATE ION | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Enzymatic Off-Loading of Hybrid Polyketides by Dieckmann Condensation.
Acs Chem.Biol., 2020
|
|
7X2K
 
 | | Crystal structure of nanobody Nb70 with antibody 1F11 fab and SARS-CoV-2 RBD | | Descriptor: | 1F11-H, 1F11-L, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
5FPL
 
 | | Crystal structure of human JARID1B in complex with CCT363901 | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-(2-azanylethyl)pyrazol-1-yl]-3H-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Yann-Vai, L.B, Nowak, R, Johansson, C, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Huber, K, Oppermann, U. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 8-Substituted Pyrido[3,4-D]Pyrimidin-4(3H)-One Derivatives as Potent, Cell Permeable, Kdm4 (Jmjd2) and Kdm5 (Jarid1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
9IIJ
 
 | |
8T2J
 
 | | Structure of the catalytic domain of PPM1D/Wip1 serine/threonine phosphatase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumar, J.P, Kosek, D, Dyda, F. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanistic studies of the PPM1D serine/threonine phosphatase catalytic domain.
J.Biol.Chem., 300, 2024
|
|
8ENW
 
 | |
5NQC
 
 | | CK2alpha in complex with NMR154 | | Descriptor: | (3E)-6,7-dichloro-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-one, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Seetoh, W.-G, Stubbs, C.J. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disrupting the CK2alpha-CK2beta protein-protein interaction within the protein kinase CK2 heterotetramer using a fragment-based approach
To Be Published
|
|
6EJ4
 
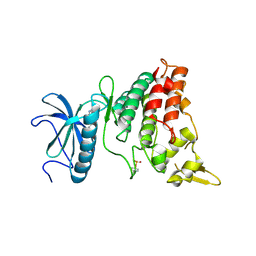 | | DYRK1A in complex with XMD7-112 | | Descriptor: | 3-(3-pyridin-3-yl-1~{H}-pyrrolo[2,3-b]pyridin-5-yl)aniline, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
8EJV
 
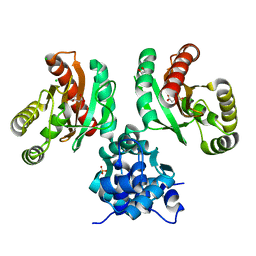 | | The crystal structure of Pseudomonas putida PcaR in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Functional and structural characterization of an IclR family transcription factor for the development of dicarboxylic acid biosensors.
Febs J., 291, 2024
|
|
6IES
 
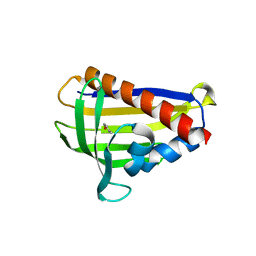 | | Onion lachrymatory factor synthase (LFS) containing (E)-2-propen 1-ol (crotyl alcohol) | | Descriptor: | (2E)-but-2-en-1-ol, Lachrymatory-factor synthase | | Authors: | Sato, Y, Arakawa, T, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase.
Acs Catalysis, 10, 2020
|
|
6P88
 
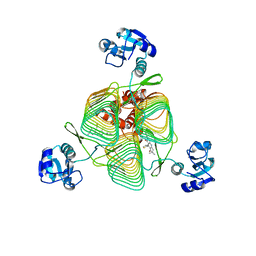 | | E.coli LpxD in complex with compound 6 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-{3-[(furan-2-carbonyl)amino]phenyl}-2,3-dihydro-1,4-benzodioxine-6-carboxamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6I71
 
 | |
6IJR
 
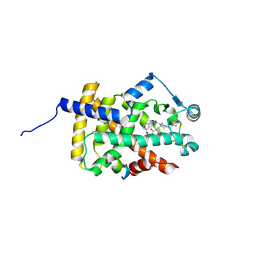 | | Human PPARgamma ligand binding domain complexed with SB1495 | | Descriptor: | 16 mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1S,2R)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6I7I
 
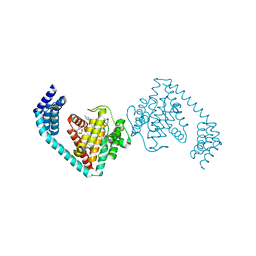 | | Crystal structure of dimeric FICD mutant K256A complexed with MgATP | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
