5UIK
 
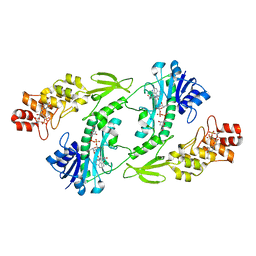 | | X-ray structure of the FdtF formyltransferase from salmonella enteric O60 in complex with TDP-Fuc3N and folinic acid | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, Formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
5V6U
 
 | |
5VPK
 
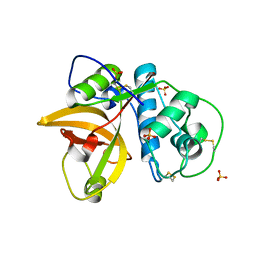 | | CRYSTAL STRUCTURE OF MITE ALLERGEN DER F 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Der f 1 variant, SULFATE ION | | Authors: | Chruszcz, M, Chapman, M.D, Vailes, L.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Of Mite Allergens Der F 1 And Der P 1 Reveal Differences In Surface-Exposed Residues That May Influence Antibody Binding
J.Mol.Biol., 386, 2009
|
|
5VXS
 
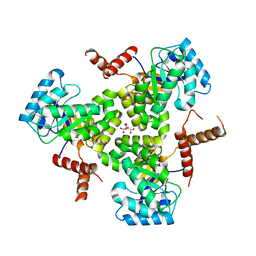 | | Crystal Structure Analysis of human CLYBL in apo form | | Descriptor: | CITRIC ACID, Citrate lyase subunit beta-like protein, mitochondrial | | Authors: | Shen, H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | The Human Knockout Gene CLYBL Connects Itaconate to Vitamin B12.
Cell, 171, 2017
|
|
5T4O
 
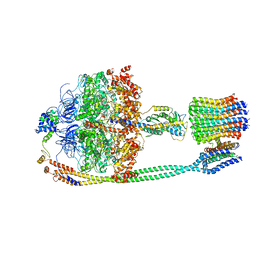 | | Autoinhibited E. coli ATP synthase state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
5T4M
 
 | |
2HD0
 
 | |
2H9H
 
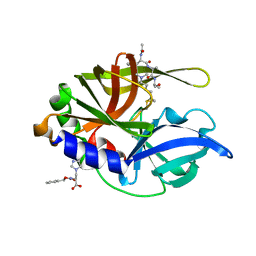 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A virus protease 3C, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Three residue peptide | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-09 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
5T4Q
 
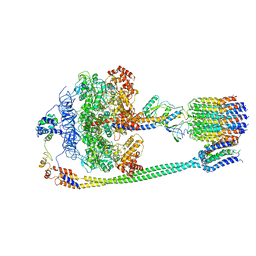 | | Autoinhibited E. coli ATP synthase state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.53 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
2HRV
 
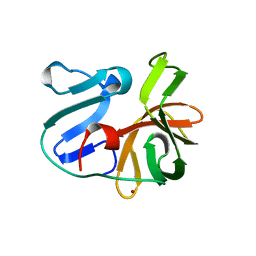 | | 2A CYSTEINE PROTEINASE FROM HUMAN RHINOVIRUS 2 | | Descriptor: | 2A CYSTEINE PROTEINASE, ZINC ION | | Authors: | Petersen, J.F.W, Cherney, M.M, Liebig, H.-D, Skern, T, Kuechler, E, James, M.N.G. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis.
EMBO J., 18, 1999
|
|
5VSG
 
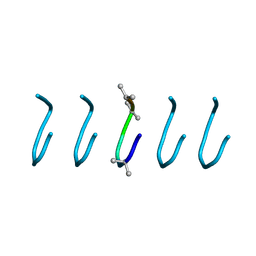 | | Fibrils of the super helical repeat peptide, SHR-FF, grown at elevated temperature | | Descriptor: | Super Helical Repeat Peptide SHR-FF | | Authors: | Mondal, S, Sawaya, M.R, Eisenberg, D.S, Gazit, E. | | Deposit date: | 2017-05-11 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Transition of Metastable Cross-alpha Crystals into Cross-beta Fibrils by beta-Turn Flipping.
J.Am.Chem.Soc., 141, 2019
|
|
5VXC
 
 | |
2HAL
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A Protease 3C, N-ACETYL-LEUCYL-PHENYLALANYL-PHENYLALANYL-GLUTAMATE-FLUOROMETHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
5W62
 
 | | Crystal structure of mouse BAX monomer | | Descriptor: | Apoptosis regulator BAX, SULFATE ION | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E, Luo, C.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
5VKI
 
 | | Crystal structure of P[19] rotavirus VP8* complexed with mucin core 2 | | Descriptor: | GLYCEROL, Outer capsid protein VP4, SULFATE ION, ... | | Authors: | Xu, S, Liu, Y, Woodruff, A, Zhong, W, Jiang, X, Kennedy, M.A. | | Deposit date: | 2017-04-21 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of glycan specificity of P[19] VP8*: Implications for rotavirus zoonosis and evolution.
PLoS Pathog., 13, 2017
|
|
5VKS
 
 | | Crystal structure of P[19] rotavirus VP8* complexed with LNFPI | | Descriptor: | GLYCEROL, Outer capsid protein VP4, SULFATE ION, ... | | Authors: | Xu, S, Liu, Y, Woodruff, A, Zhong, W, Jiang, X, Kennedy, M.A. | | Deposit date: | 2017-04-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis of glycan specificity of P[19] VP8*: Implications for rotavirus zoonosis and evolution.
PLoS Pathog., 13, 2017
|
|
2HG4
 
 | | Structure of the ketosynthase-acyltransferase didomain of module 5 from DEBS. | | Descriptor: | 6-Deoxyerythronolide B Synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tang, Y, Kim, C.Y, Mathews, I.I, Cane, D.E, Khosla, C. | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The 2.7-A crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HPP
 
 | |
5T4N
 
 | |
5W5X
 
 | | Crystal structure of BAXP168G in complex with an activating antibody | | Descriptor: | 1,2-ETHANEDIOL, 3C10 Fab' heavy chain, 3C10 Fab' light chain, ... | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
2LNC
 
 | | Solution NMR structure of Norwalk virus protease | | Descriptor: | 3C-like protease | | Authors: | Takahashi, D, Hiromasa, Y, Kim, Y, Anbanandam, A, Chang, K, Prakash, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics characterization of norovirus protease.
Protein Sci., 22, 2013
|
|
5W1W
 
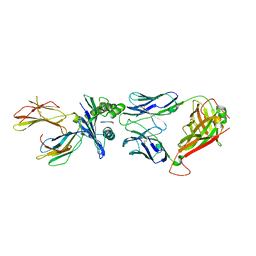 | | Structure of the HLA-E-VMAPRTLVL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-10-04 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
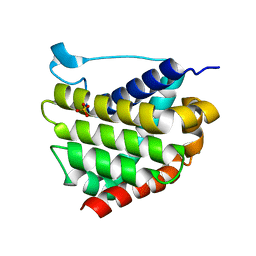
5W60
 
 | |
5W61
 
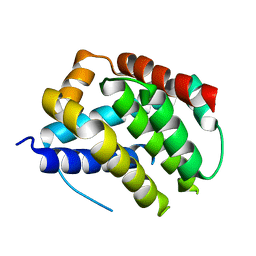 | |
5W3N
 
 | | Molecular structure of FUS low sequence complexity domain protein fibrils | | Descriptor: | RNA-binding protein FUS | | Authors: | Murray, D.T, Kato, M, Lin, Y, Thurber, K, Hung, I, McKnight, S, Tycko, R. | | Deposit date: | 2017-06-08 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains.
Cell, 171, 2017
|
|
