1AZS
 
 | |
5O2T
 
 | | Human KRAS in complex with darpin K27 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange.
Nat Commun, 8, 2017
|
|
2BTR
 
 | | STRUCTURE OF CDK2 COMPLEXED WITH PNU-198873 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(5-ISOPROPYL-THIAZOL-2-YL)-2-PYRIDIN-3-YL-ACETAMIDE | | Authors: | Vulpetti, A, Casale, E, Roletto, F, Amici, R, Villa, M, Pevarello, P. | | Deposit date: | 2005-06-06 | | Release date: | 2005-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Drug Design to the Discovery of New 2-Aminothiazole Cdk2 Inhibitors.
J.Mol.Graph.Model., 24, 2006
|
|
7B1P
 
 | | Crystal Structure of Human BACE-1 in Complex with Compound 38a (NB-854) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-25 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7B1Q
 
 | | Crystal Structure of Human BACE-1 in Complex with Compound NB-360 (compound 54) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]-4-fluoranyl-phenyl]-5-cyano-3-methyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-25 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7B1E
 
 | | BACE1 IN COMPLEX WITH compound 3 (NB-641) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(4~{S})-2-azanyl-4-methyl-5,6-dihydro-1,3-thiazin-4-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
1W6J
 
 | | Structure of human OSC in complex with Ro 48-8071 | | Descriptor: | LANOSTEROL SYNTHASE, TETRADECANE, [4-({6-[ALLYL(METHYL)AMINO]HEXYL}OXY)-2-FLUOROPHENYL](4-BROMOPHENYL)METHANONE, ... | | Authors: | Thoma, R, Schulz-Gasch, T, D'Arcy, B, Benz, J, Aebi, J, Dehmlow, H, Hennig, M, Ruf, A. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insight Into Steroid Scaffold Formation from the Structure of Human Oxidosqualene Cyclase
Nature, 432, 2004
|
|
1W6K
 
 | | Structure of human OSC in complex with Lanosterol | | Descriptor: | LANOSTEROL, LANOSTEROL SYNTHASE, octyl beta-D-glucopyranoside | | Authors: | Thoma, R, Schulz-Gasch, T, D'Arcy, B, Benz, J, Aebi, J, Dehmlow, H, Hennig, M, Ruf, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insight Into Steroid Scaffold Formation from the Structure of Human Oxidosqualene Cyclase
Nature, 432, 2004
|
|
2AIX
 
 | |
2N3P
 
 | |
2N2G
 
 | |
1CHV
 
 | |
2PYY
 
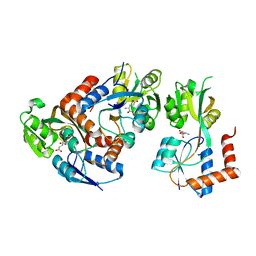 | | Crystal Structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with (L)-glutamate | | Descriptor: | GLUTAMIC ACID, Ionotropic glutamate receptor bacterial homologue | | Authors: | Lee, J.H, Kang, G.B, Lim, H.-H, Ree, M, Park, C.-S, Eom, S.H. | | Deposit date: | 2007-05-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with L-glutamate: structural dissection of the ligand interaction and subunit interface.
J.Mol.Biol., 376, 2008
|
|
2M6X
 
 | |
3EJF
 
 | |
4P6A
 
 | | Crystal structure of a potent anti-HIV lectin actinohivin in complex with alpha-1,2-mannotriose | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Zhang, F, Hoque, M.M, Suzuki, K, Tsunoda, M, Naomi, O, Tanaka, H, Takenaka, A. | | Deposit date: | 2014-03-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | The characteristic structure of anti-HIV actinohivin in complex with three HMTG D1 chains of HIV-gp120.
Chembiochem, 15, 2014
|
|
3EKE
 
 | | Crystal structure of IBV X-domain at pH 5.6 | | Descriptor: | L(+)-TARTARIC ACID, Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
5I2K
 
 | | Structure of the human GluN1/GluN2A LBD in complex with 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 19) | | Descriptor: | 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
3EJG
 
 | | Crystal structure of HCoV-229E X-domain | | Descriptor: | Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-18 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
3CTK
 
 | | Crystal structure of the type 1 RIP bouganin | | Descriptor: | rRNA N-glycosidase | | Authors: | Fermani, S, Tosi, G, Falini, G, Ripamonti, A, Farini, V, Bolognesi, A, Polito, L. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure/function studies on two type 1 ribosome inactivating proteins: Bouganin and lychnin.
J.Struct.Biol., 168, 2009
|
|
4W1V
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a thiazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, CHLORIDE ION, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Target-Based Identification of Whole-Cell Active Inhibitors of Biotin Biosynthesis in Mycobacterium tuberculosis.
Chem.Biol., 22, 2015
|
|
6PGZ
 
 | | MTSL labelled T4 lysozyme pseudo-wild type V75C mutant | | Descriptor: | CHLORIDE ION, Endolysin, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
2D43
 
 | | Crystal structure of arabinofuranosidase complexed with arabinotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Miwa, Y, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-10-07 | | Release date: | 2006-09-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The family 42 carbohydrate-binding module of family 54 alpha-L-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose
Biochem.J., 399, 2006
|
|
4W1X
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with 1-(4-(4-(3-chlorobenzoyl)piperazin-1-yl)phenyl)ethanone | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-(3-chlorobenzoyl)piperazin-1-yl]phenyl}ethanone, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target-Based Identification of Whole-Cell Active Inhibitors of Biotin Biosynthesis in Mycobacterium tuberculosis.
Chem.Biol., 22, 2015
|
|
1U65
 
 | | Ache W. CPT-11 | | Descriptor: | (4S)-4,11-DIETHYL-4-HYDROXY-3,14-DIOXO-3,4,12,14-TETRAHYDRO-1H-PYRANO[3',4':6,7]INDOLIZINO[1,2-B]QUINOLIN-9-YL 1,4'-BIPIPERIDINE-1'-CARBOXYLATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Harel, M, Hyatt, J.L, Brumshtein, B, Morton, C.L, Wadkins, R.W, Silman, I, Sussman, J.L, Potter, P.M, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-07-29 | | Release date: | 2005-07-19 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The crystal structure of the complex of the anticancer prodrug 7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothecin (CPT-11) with Torpedo californica acetylcholinesterase provides a molecular explanation for its cholinergic action
Mol.Pharmacol., 67, 2005
|
|
