7OYT
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88L in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
8OJM
 
 | | Galectin-3 in complex with 2,6-anhydro-3-deoxy-3-S-(beta-D-galactopyranosyl)-3-thio-D-glycero-D-galacto-heptonamide | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxane-2-carboxamide, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
6G36
 
 | | Crystal structure of haspin in complex with 5-chlorotubercidin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chlorotubercidin, COBALT (II) ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5IDY
 
 | |
7RKK
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (C2 space group) | | Descriptor: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein | | Authors: | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5IZ3
 
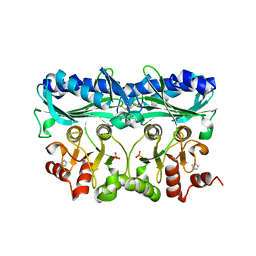 | | P. patens sedoheptulose-1,7-bisphosphatase | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, Predicted protein, ... | | Authors: | Einsle, O, Guetle, D. | | Deposit date: | 2016-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ZXN
 
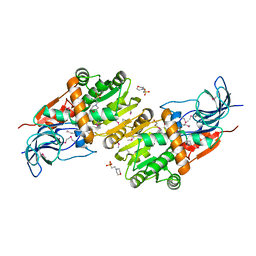 | | Crystal structure of CurA from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP-dependent oxidoreductase | | Authors: | Kim, M.-K, Bae, D.-W, Cha, S.-S. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Structural and Biochemical Characterization of the Curcumin-Reducing Activity of CurA from Vibrio vulnificus.
J. Agric. Food Chem., 66, 2018
|
|
4QF7
 
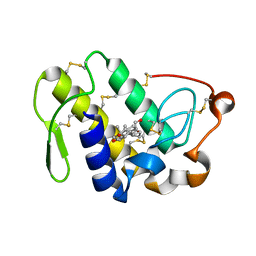 | | Crystal Structure of the Complex of Phospholipase A2 with Corticosterone at 1.48 A Resolution | | Descriptor: | CORTICOSTERONE, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-20 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structures and binding studies of the complexes of phospholipase A2 with five inhibitors
Biochim.Biophys.Acta, 1854, 2015
|
|
6XOO
 
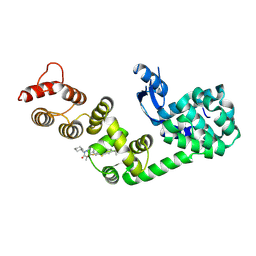 | | DCN1 bound to DI-1859 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-{(1S)-1-cyclohexyl-2-[(2-methylpropanoyl)amino]ethyl}-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOM
 
 | | DCN1 bound to 8 | | Descriptor: | (2R)-N-[(2S)-2-cyclohexyl-2-({N-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alanyl}amino)ethyl]-2-methyl-4-(morpholin-4-yl)butanamide, 1,2-ETHANEDIOL, Lysozyme, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
8U5P
 
 | | Structure of Mango II aptamer bound to T01-6A-B | | Descriptor: | 3-[2,16-dioxo-20-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-6,9,12-trioxa-3,15-diazaicosan-1-yl]-2-{(E)-[6-(4-methoxyphenyl)-1-methylquinolin-4(1H)-ylidene]methyl}-1,3-benzothiazol-3-ium, Mango II, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Symmetry breaking of fluorophore binding to a G-quadruplex generates an RNA aptamer with picomolar KD.
Nucleic Acids Res., 52, 2024
|
|
8U60
 
 | | Structure of Mango II variant2 aptamer bound to T01-6A | | Descriptor: | 2-[(~{E})-[6-(4-methoxyphenyl)-1-methyl-quinolin-4-ylidene]methyl]-3-methyl-1,3-benzothiazole, POTASSIUM ION, RNA Mango II variant 2 aptamer | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-27 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Symmetry breaking of fluorophore binding to a G-quadruplex generates an RNA aptamer with picomolar KD.
Nucleic Acids Res., 52, 2024
|
|
8OJK
 
 | | Galectin-3 in complex with 2,6-anhydro-3-deoxy-3-S-(beta-D-galactopyranosyl)-3-thio-D-glycero-L-altro-heptonamide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxane-2-carboxamide, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
6XOL
 
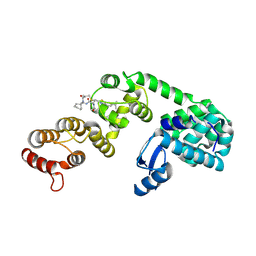 | | DCN1 bound to DI-1548 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-{(1S)-1-cyclohexyl-2-[(2-methylpropanoyl)amino]ethyl}-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
8U5K
 
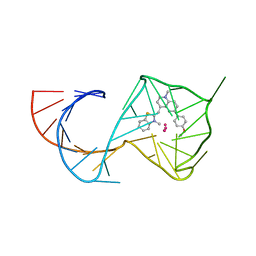 | | Structure of Mango II aptamer bound to T01-6A | | Descriptor: | 2-[(~{E})-[6-(4-methoxyphenyl)-1-methyl-quinolin-4-ylidene]methyl]-3-methyl-1,3-benzothiazole, Mango II, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Symmetry breaking of fluorophore binding to a G-quadruplex generates an RNA aptamer with picomolar KD.
Nucleic Acids Res., 52, 2024
|
|
6PGY
 
 | |
9JOX
 
 | |
6XAK
 
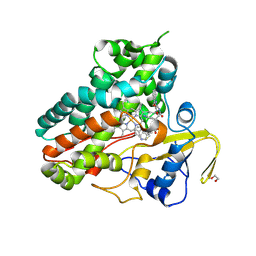 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-Pro) and cyclo-(L-Trp-L-Trp) | | Descriptor: | (3S,6S)-3,6-bis[(1H-indol-3-yl)methyl]piperazine-2,5-dione, (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
9FXG
 
 | | Crystal structure of double mutant Y115E Y117E human Glutaminyl Cyclase in apo-state | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective.
Int J Mol Sci, 25, 2024
|
|
8IPC
 
 | |
6PLJ
 
 | | A nucleotidyl transferase from Methanothermobacter thermautotroptrophicus (Mth528) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Carbone, V, Schofield, L, Sutherland-Smith, A, Ronimus, R. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A nucleotidyl transferase from Methanothermobacter thermautotroptrophicus (Mth528)
To Be Published
|
|
6XIN
 
 | | The crystal structure of tryptophan synthase from Salmonella enterica serovar typhimurium in complex with (2S)-3-Amino-3-imino-2-phenyldiazenylpropanamide at the enzyme alpha-site. | | Descriptor: | (2R,3Z)-3-amino-3-imino-2-[(E)-phenyldiazenyl]propanamide, 1,2-ETHANEDIOL, CESIUM ION, ... | | Authors: | Hilario, E, Chang, C, Mueller, L.J, Dunn, M.F, Fan, L. | | Deposit date: | 2020-06-20 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of tryptophan synthase from Salmonella enterica serovar typhimurium in complex with (2S)-3-Amino-3-imino-2-phenyldiazenylpropanamide at the enzyme alpha-site.
To Be Published
|
|
5MW6
 
 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
8U4W
 
 | |
5J20
 
 | | HSP90 in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-N-furan-2-ylmethyl-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(furan-2-yl)methyl]-2,4-dihydroxy-N-methylbenzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
