6R9U
 
 | | Human Cyclophilin D in complex with fragment | | Descriptor: | 14-ethyl-4,6-dioxa-10,14-diazatricyclo[7.6.0.0^{3,7}]pentadeca-1(9),2,7-trien-13-one, Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ... | | Authors: | Graedler, U. | | Deposit date: | 2019-04-04 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8KB2
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - iodide derivative | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8H4Q
 
 | | Aspergillomarasmine A biosynthese complex with OPS | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Rhodanese domain-containing protein | | Authors: | Lu, M, Zhang, J, Wang, Z, Han, L. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Investigation of AMA Synthase from Pyrenophora teres f. teres 0-1 for the Synthesis of Aspergillomarasmine A Analogues and Non-Canonical Amino Acids
Adv.Synth.Catal., 366, 2024
|
|
8H7N
 
 | | Structure of nanobody 11A in complex with triazophos | | Descriptor: | 1,2-ETHANEDIOL, Nanobody 11A, SODIUM ION, ... | | Authors: | Wang, H, Li, J.D, Shen, X, Xu, Z.L, Sun, Y.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Insights into the Stability and Recognition Mechanism of the Antiquinalphos Nanobody for the Detection of Quinalphos in Foods.
Anal.Chem., 95, 2023
|
|
6Z5E
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004093 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 11-cyclopropyl-14-(2-hydroxyethyl)-8,11,14,18,19,22-hexazatetracyclo[13.5.2.12,6.018,21]tricosa-1(21),2(23),3,5,15(22),16,19-heptaen-7-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004093
To Be Published
|
|
8H4H
 
 | | The apo structure of Aspergillomarasmine A synthetase | | Descriptor: | Rhodanese domain-containing protein | | Authors: | Lu, M, Zhang, J, Wang, Z, Han, L. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of AMA Synthase from Pyrenophora teres f. teres 0-1 for the Synthesis of Aspergillomarasmine A Analogues and Non-Canonical Amino Acids
Adv.Synth.Catal., 366, 2024
|
|
9HY5
 
 | | sc-4E (scFv derived from the mAb 4E1 against CD93) crystallized at pH 9.0 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Raucci, L, Orlandini, M, Pozzi, C. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and antigen-binding surface definition of an anti-CD93 monoclonal antibody for the treatment of degenerative vascular eye diseases.
Int.J.Biol.Macromol., 309, 2025
|
|
6E1V
 
 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 3: 2-[(9H-carbazol-3-yl)oxy]-N,N-dimethylethan-1-amine | | Descriptor: | 2-[(9H-carbazol-3-yl)oxy]-N,N-dimethylethan-1-amine, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
9HY2
 
 | | sc-4E (scFv derived from the mAb 4E1 against CD93) crystallized at pH 7.5 (monomeric state from gel filtration) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Raucci, L, Orlandini, M, Pozzi, C. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and antigen-binding surface definition of an anti-CD93 monoclonal antibody for the treatment of degenerative vascular eye diseases.
Int.J.Biol.Macromol., 309, 2025
|
|
6IUO
 
 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
8EJO
 
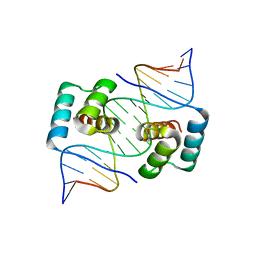 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EJP
 
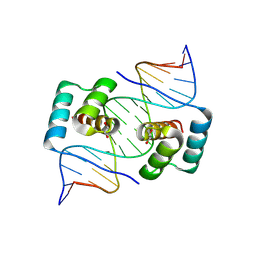 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
6YPG
 
 | |
9BS1
 
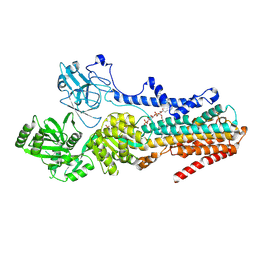 | | Cryo-EM structure of the S. cerevisiae lipid flippase Neo1 bound with PI4P in the E2P state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl di[(9Z)-octadec-9-enoate], BERYLLIUM TRIFLUORIDE ION, Phospholipid-transporting ATPase NEO1 | | Authors: | Duan, H.D, Li, H. | | Deposit date: | 2024-05-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | P4-ATPases control phosphoinositide membrane asymmetry and neomycin resistance.
Nat.Cell Biol., 27, 2025
|
|
8H7X
 
 | | Crystal structure of EGFR T790M/C797S mutant in complex with brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | A macrocyclic kinase inhibitor overcomes triple resistant mutations in EGFR-positive lung cancer.
NPJ Precis Oncol, 8, 2024
|
|
9B88
 
 | | Anti-OspC Fab 8C1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Heavy Chain of monoclonal 8C1 Fab, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2024-03-29 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of anti-OspC Fab 8C1.
To Be Published
|
|
5LTD
 
 | | Phosphate-bound Pichia angusta Atg18 | | Descriptor: | 1,2-ETHANEDIOL, Autophagy-related protein 18, PHOSPHATE ION | | Authors: | Scacioc, A, Kuhnel, K. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phopshate-bound Pichia angusta Atg18
To Be Published
|
|
8Y56
 
 | | Cryo-EM reveals cholesterol binding in the lysosomal GPCR-like protein LYCHOS | | Descriptor: | CHOLESTEROL, Lysosomal cholesterol signaling protein, SODIUM ION, ... | | Authors: | Zhao, J, Shen, Q.Y, Zhang, Y, Shao, Z.H. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM reveals cholesterol binding in the lysosomal GPCR-like protein LYCHOS.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8ER5
 
 | | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NlpC/P60 domain-containing protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448.
To Be Published
|
|
6FRC
 
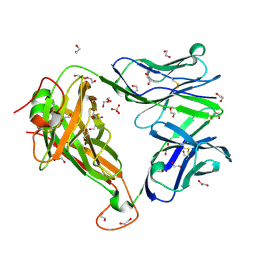 | |
4U0V
 
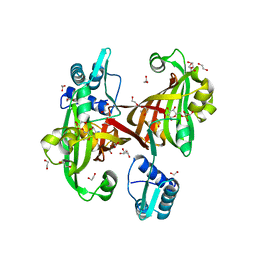 | |
9J5A
 
 | |
6RTW
 
 | | Crystal structure of the Patched-1 (PTCH1) ectodomain in complex with nanobody NB64 and cholesterol-hemisuccinate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Llama-derived nanobody NB64, ... | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
5M0W
 
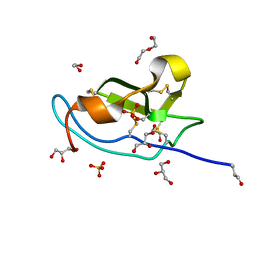 | | N-terminal domain of mouse Shisa 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lohkamp, B, Ojala, J.R.M. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ab initio solution of macromolecular crystal structures without direct methods.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5L83
 
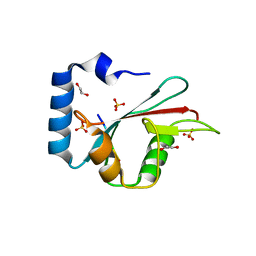 | | Complex of potato ATG8 protein with a peptide from Irish potato famine pathogen effector protein PexRD54 | | Descriptor: | 1,2-ETHANEDIOL, ASP-TRP-GLU-ILE-VAL, Autophagy-related protein, ... | | Authors: | Maqbool, A, Hughes, R.K, Banfield, M.J. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Host Autophagy-related Protein 8 (ATG8) Binding by the Irish Potato Famine Pathogen Effector Protein PexRD54.
J.Biol.Chem., 291, 2016
|
|
