8DPQ
 
 | | Beta-lactamase CTX-M-14 N170A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Lu, S, Neetu, N, Palzkill, T. | | Deposit date: | 2022-07-15 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
6QE4
 
 | | Re-refinement of 5OLI human IBA57-I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Putative transferase CAF17, mitochondrial | | Authors: | Calderone, V, Ciofi-Baffoni, S, Gourdoupis, S, Banci, L, Nasta, V. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In-house high-energy-remote SAD phasing using the magic triangle: how to tackle the P1 low symmetry using multiple orientations of the same crystal of human IBA57 to increase the multiplicity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4R7S
 
 | |
5MO2
 
 | | Neutron structure of cationic trypsin in complex with N-amidinopiperidine | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-13 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
9B3U
 
 | | Rat TRPV2 WT bound to probenecid | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 4-(dipropylsulfamoyl)benzoic acid, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Rocereta, J.A, Pumroy, R.A, Moiseenkova-Bell, V.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural insights into TRPV2 modulation by probenecid.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7S71
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI35 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(hexylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
9B3W
 
 | | Rat TRPV2 E724A/D725A Apo | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Pumroy, R.A, Rocereta, J.A, Moiseenkova-Bell, V.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into TRPV2 modulation by probenecid.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7S72
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI36 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
4QQ8
 
 | | Crystal structure of the formolase FLS in space group P 43 21 2 | | Descriptor: | 1,2-ETHANEDIOL, Formolase, MAGNESIUM ION, ... | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L. | | Deposit date: | 2014-06-26 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7S6W
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI29 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6X
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI30 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S75
 
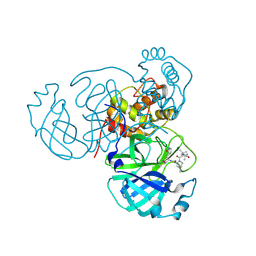 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI42 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(3-methylbutanoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6Y
 
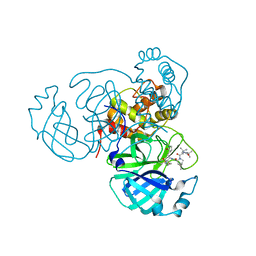 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI32 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-[(cyclopropylmethyl)amino]-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
8BGN
 
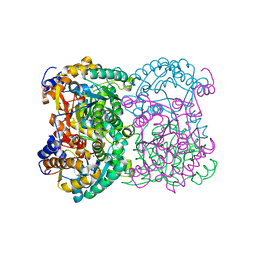 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Diacetylchitobiose deacetylase, ... | | Authors: | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
7S70
 
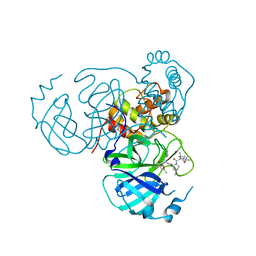 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI34 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(butylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
5MQT
 
 | | Crystal structure of dCK mutant C3S in complex with imatinib and UDP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Deoxycytidine kinase, ... | | Authors: | Saez-Ayala, M, Rebuffet, E, Hammam, K, Gros, L, Lopez, S, Hajem, B, Humbert, M, Baudelet, E, Audebert, S, Betzi, S, Lugari, A, Combes, S, Pez, D, Letard, S, Mansfield, C, Moussy, A, de Sepulveda, P, Morelli, X, Dubreuil, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology.
Nat Commun, 8, 2017
|
|
9B3V
 
 | |
7S6Z
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
6NJJ
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN14770 | | Descriptor: | (4-{[2-(3-chlorophenyl)-6-(trifluoromethyl)pyridin-4-yl]methyl}phenyl)acetic acid, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2019-01-03 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders.
J.Med.Chem., 62, 2019
|
|
8GMH
 
 | | Crystal Structure of the ternary complex of TelA-LXG, LapA3, and LapA4 | | Descriptor: | 1,2-ETHANEDIOL, LXG domain-containing protein, LapA3, ... | | Authors: | Klein, T.A, Shah, P.Y, Gkragkopoulou, P, Grebenc, D.W, Kim, Y, Whitney, J.C. | | Deposit date: | 2023-03-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a tripartite protein complex that targets toxins to the type VII secretion system.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
7PZS
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2S,4S)-N-((3R,5R)-5-((2-chloro-4-((4-(morpholinomethyl)phenyl)ethynyl)benzamido)methyl)-1-(cyclopropanecarbonyl)pyrrolidin-3-yl)-4-fluoropyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
6H09
 
 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2018-07-06 | | Release date: | 2018-08-15 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
7Z8E
 
 | | Crystal structure of the substrate-binding protein YejA from S. meliloti in complex with peptide fragment | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, GLY-SER-ASP-VAL-ALA, ... | | Authors: | Morera, S, Vigouroux, V, Travin, D.Y. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual-Uptake Mode of the Antibiotic Phazolicin Prevents Resistance Acquisition by Gram-Negative Bacteria.
Mbio, 14, 2023
|
|
8B9M
 
 | | Cysteine Synthase from Leishmania Infantum | | Descriptor: | CHLORIDE ION, Cysteine synthase, GLYCEROL | | Authors: | Sowerby, K.V, Pohl, E. | | Deposit date: | 2022-10-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
