3MRL
 
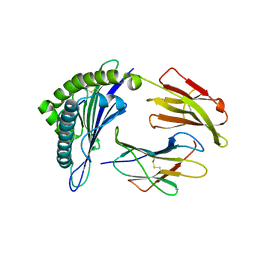 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide C6V variant | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Reiser, J.-B, Chouquet, A, Le Gorrec, M, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
3NNU
 
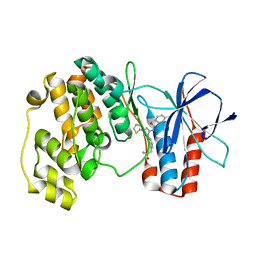 | | Crystal structure of P38 alpha in complex with DP1376 | | Descriptor: | 2-{3-[(5E)-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-3-thiophen-2-yl-2,5-dihydro-1H-pyrazol-1-yl]phenyl}acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNW
 
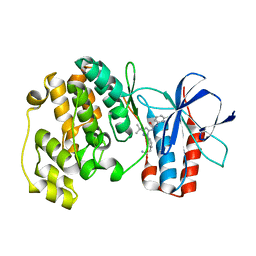 | | Crystal structure of P38 alpha in complex with DP802 | | Descriptor: | 2-[3-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-2,5-dihydro-1H-pyrazol-1-yl)phenyl]acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNV
 
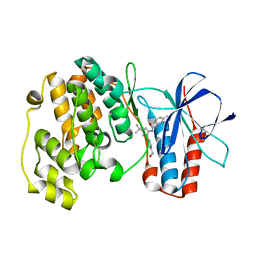 | | Crystal structure of P38 alpha in complex with DP437 | | Descriptor: | 1-{3-tert-butyl-1-[4-(hydroxymethyl)phenyl]-1H-pyrazol-5-yl}-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OOJ
 
 | |
8BDZ
 
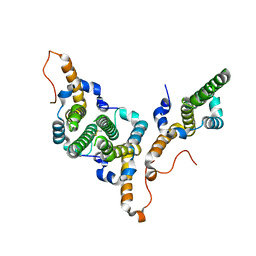 | | Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) with T=4 topology | | Descriptor: | Core protein,Matrix protein 2,External core antigen | | Authors: | Egorov, V.V, Shvetsov, A.V, Pichkur, E.B, Shaldzhyan, A.A, Zabrodskaya, Y.A, Vinogradova, D.S, Nekrasov, P.A, Gorshkov, A.N, Garmay, Y.P, Kovaleva, A.A, Stepanova, L.A, Tsybalova, L.M, Shtam, T.A, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2022-10-20 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Inside and outside of virus-like particles HBc and HBc/4M2e: A comprehensive study of the structure.
Biophys.Chem., 293, 2022
|
|
3MRG
 
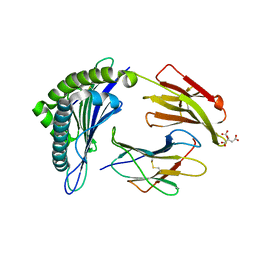 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Gras, S, Reiser, J.-B, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
4DME
 
 | | GCN4 leucine zipper domain in a trimeric oligomerization state | | Descriptor: | GCN4-p1 leucine zipper domain, SULFATE ION | | Authors: | Oshaben, K.M, Salari, R, Chong, L.T, Horne, W.S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Native GCN4 Leucine-Zipper Domain Does Not Uniquely Specify a Dimeric Oligomerization State.
Biochemistry, 51, 2012
|
|
4E22
 
 | |
3MRH
 
 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide N3S variant | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Reiser, J.-B, Chouquet, A, Le Gorrec, M, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
5C03
 
 | | Crystal Structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Min, X, Wang, Z, Walker, N. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the JH2 Pseudokinase Domain of JAK Family Tyrosine Kinase 2 (TYK2).
J.Biol.Chem., 290, 2015
|
|
4QF9
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid at 2.28 A resolution | | Descriptor: | (2S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kristensen, C.M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2014-05-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Binding Mode of an alpha-Amino Acid-Linked Quinoxaline-2,3-dione Analogue at Glutamate Receptor Subtype GluK1.
ACS Chem Neurosci, 6, 2015
|
|
5CMM
 
 | |
6SL1
 
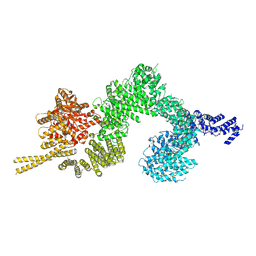 | | Structure of the open conformation of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
7YBJ
 
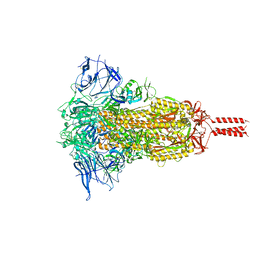 | | SARS-CoV-2 Mu variant spike(close state) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBI
 
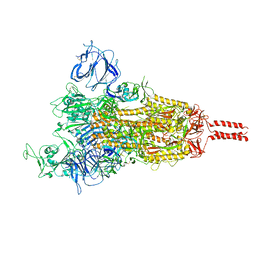 | | SARS-CoV-2 Mu variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
6SKZ
 
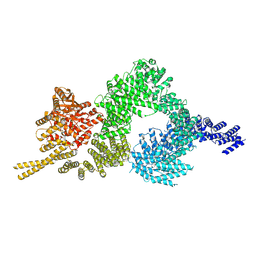 | | Structure of the closed conformation of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
5CMK
 
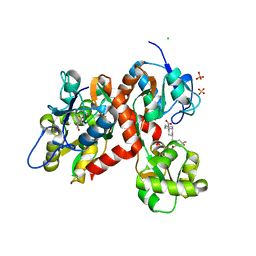 | | Crystal structure of the GluK2EM LBD dimer assembly complex with glutamate and LY466195 | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Chittori, S, Mayer, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
4DMD
 
 | |
7YBL
 
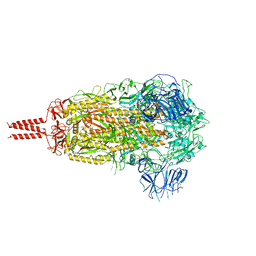 | | SARS-CoV-2 B.1.620 variant spike (close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBH
 
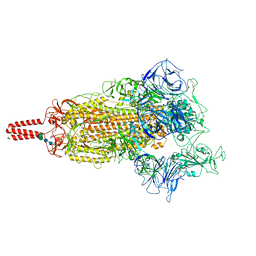 | | SARS-CoV-2 lambda variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBK
 
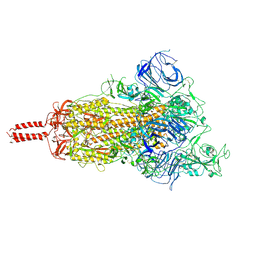 | | SARS-CoV-2 B.1.620 variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBM
 
 | | SARS-CoV-2 C.1.2 variant spike (Close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBN
 
 | | SARS-CoV-2 C.1.2 variant spike (Open state) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
6SKY
 
 | | FAT and kinase domain of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
