7PZW
 
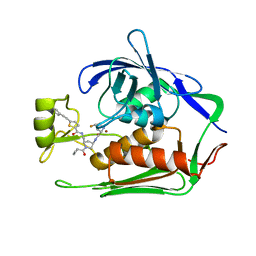 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (4S)-N-((3R,5R)-1-(cyclopropanecarbonyl)-5-((5-(phenylethynyl)thiophene-2-carboxamido)methyl)pyrrolidin-3-yl)-4-fluoropyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
8VIS
 
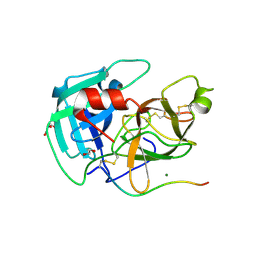 | | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fraser, B.J, Dong, A, Ilyassov, O, Kenney, T, Li, Y.Y, Seitova, A, Li, Y, Hejazi, Z, Edwards, A, Benard, F, Arrowsmith, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-01-31 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of TMPRSS11D specificity and autocleavage activation.
Nat Commun, 16, 2025
|
|
6WHY
 
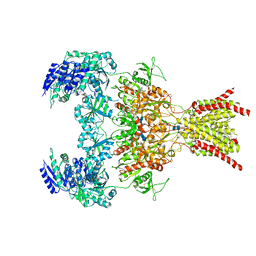 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
8VR7
 
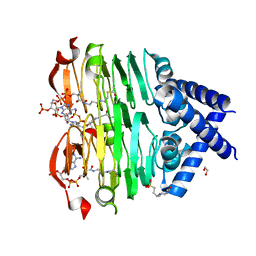 | | crystal structure of the Pcryo_0619 N-acetyltransferase from Psychrobacter cryohalolentis K5 int he presence of acetyl coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dunsirn, M.M, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8K5X
 
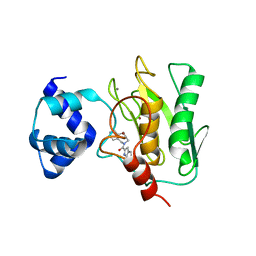 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (6-cyclopropyl-1~{H}-indol-2-yl)-(5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)methanone, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
6WIZ
 
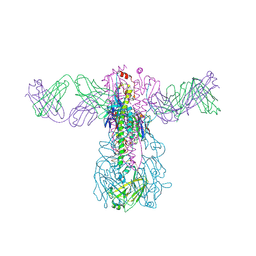 | | Crystal structure of Fab 54-1G05 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-1G05 heavy chain, Fab 54-1G05 light chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
8ZQS
 
 | |
6OP9
 
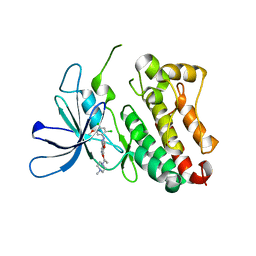 | | HER3 pseudokinase domain bound to bosutinib | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Littlefield, P, Agnew, C, Jura, N. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Targetable HER3 functions driving tumorigenic signaling in HER2-amplified cancers.
Cell Rep, 38, 2022
|
|
8VTS
 
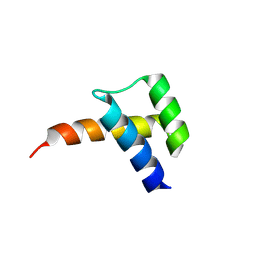 | | Meis1 homeobox domain bound to paromomycin fragment | | Descriptor: | 1,2-ETHANEDIOL, Homeobox protein Meis1, ISOPROPYL ALCOHOL, ... | | Authors: | Tomchick, D.R, Ahmed, M.S, Nguyen, N.U.N. | | Deposit date: | 2024-01-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of FDA-approved drugs that induce heart regeneration in mammals.
Nat Cardiovasc Res, 3, 2024
|
|
8K9R
 
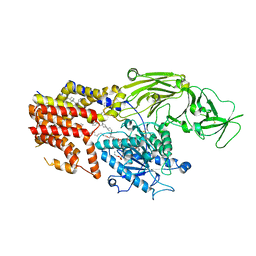 | | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8JWS
 
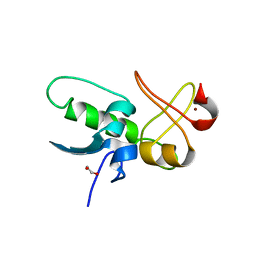 | | ePHD domain of PHD Finger Protein 7 (PHF7) | | Descriptor: | 1,2-ETHANEDIOL, PHD finger protein 7, ZINC ION | | Authors: | Bang, I, Lee, H.S, Choi, H.-J. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for PHF7-mediated ubiquitination of histone H3.
Genes Dev., 37, 2023
|
|
8K9T
 
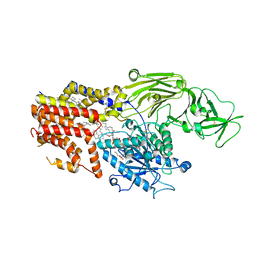 | | Cryo-EM structure of the products-bound PGAP1(Bst1)-S327A from Chaetonium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
7Q0T
 
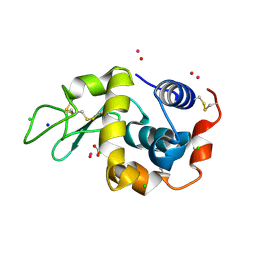 | | Lysozyme soaked with V(IV)OSO4 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Santos, M.F.A, Fernandes, A.C.P, Correia, I, Sciortino, G, Garribba, E, Santos-Silva, T, Pessoa, J.C. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Binding of V IV O 2+ , V IV OL, V IV OL 2 and V V O 2 L Moieties to Proteins: X-ray/Theoretical Characterization and Biological Implications.
Chemistry, 28, 2022
|
|
7RFW
 
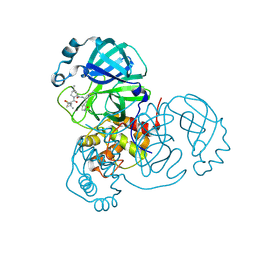 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
8B98
 
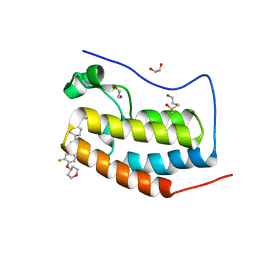 | |
8B96
 
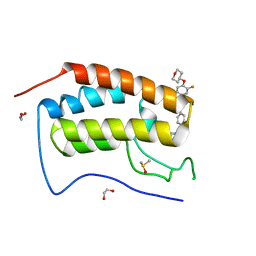 | |
7BVD
 
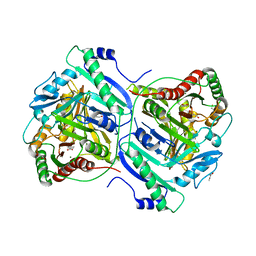 | | Anthranilate synthase component I (TrpE)[Mycolicibacterium smegmatis] | | Descriptor: | Anthranilate synthase component 1, BENZOIC ACID, GLYCEROL, ... | | Authors: | Chen, Y, Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit I of the anthranilate synthase complex of Mycolicibacterium smegmatis
Biochem.Biophys.Res.Commun., 527, 2020
|
|
7RFS
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7SWP
 
 | | G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32Q4 Fab heavy chain, G32Q4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWN
 
 | | G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32A4 Fab heavy chain, G32A4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
6O9S
 
 | |
6O9W
 
 | |
7PLN
 
 | | Structure of the APCbeta domain of Plasmodium vivax perforin-like protein 1 | | Descriptor: | Sporozoite micronemal protein essential for cell traversal, putative | | Authors: | Williams, S.I, Ni, T, Yu, X, Gilbert, R.J.C. | | Deposit date: | 2021-08-31 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural, Functional and Computational Studies of Membrane Recognition by Plasmodium Perforin-Like Proteins 1 and 2
J.Mol.Biol., 434, 2022
|
|
6WGP
 
 | | The crystal structure of a beta lactamase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a beta lactamase from Xanthomonas campestris pv. campestris str. ATCC 33913
To Be Published
|
|
8CHW
 
 | |
