8TQS
 
 | |
8OKU
 
 | | Salt-Inducible Kinase 3 in complex with an inhibitor | | Descriptor: | Serine/threonine-protein kinase SIK3, ~{N}-ethyl-4-[5-[1-(2-hydroxyethyl)pyrazol-4-yl]benzimidazol-1-yl]-2,6-dimethoxy-benzamide | | Authors: | Flower, T.G, Leonard, P.M, Lamers, M.B.A.C, Mollat, P. | | Deposit date: | 2023-03-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Optimization of Selectivity and Pharmacokinetic Properties of Salt-Inducible Kinase Inhibitors that Led to the Discovery of Pan-SIK Inhibitor GLPG3312.
J.Med.Chem., 67, 2024
|
|
5MYE
 
 | |
8U3L
 
 | | TRPV1 in nanodisc bound with empty vanilloid binding pocket at 25C | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7P4E
 
 | | Crystal structure of PPARgamma in complex with compound FL217 | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma, SULFATE ION, ... | | Authors: | Ni, X, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
6MNU
 
 | | Crystal structure of Yersinia pestis UDP-glucose pyrophosphorylase | | Descriptor: | 1,2-ETHANEDIOL, UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Gibbs, M.E, Lountos, G.T, Gumpena, R, Waugh, D.S. | | Deposit date: | 2018-10-03 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Crystal structure of UDP-glucose pyrophosphorylase from Yersinia pestis, a potential therapeutic target against plague.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8IO3
 
 | | Cryo-EM structure of human HCN3 channel with cilobradine | | Descriptor: | 3-[[(3~{S})-1-[2-(3,4-dimethoxyphenyl)ethyl]piperidin-3-yl]methyl]-7,8-dimethoxy-2,5-dihydro-1~{H}-3-benzazepin-4-one, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure of human HCN3 channel with cilobradine
To Be Published
|
|
6GUH
 
 | | CDK2 in complex with AZD5438 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
4QHT
 
 | | Crystal structure of AAA+/ sigma 54 activator domain of the flagellar regulatory protein FlrC from Vibrio cholerae in ATP analog bound state | | Descriptor: | 1,2-ETHANEDIOL, Flagellar regulatory protein C, MAGNESIUM ION, ... | | Authors: | Dey, S, Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.559 Å) | | Cite: | Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
J.Biol.Chem., 290, 2015
|
|
8FRE
 
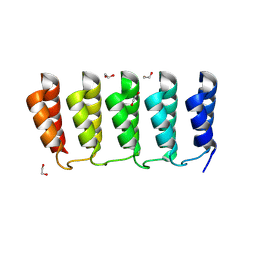 | | Designed loop protein RBL4 | | Descriptor: | 1,2-ETHANEDIOL, RBL4 | | Authors: | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | Deposit date: | 2023-01-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
7A66
 
 | |
5MEP
 
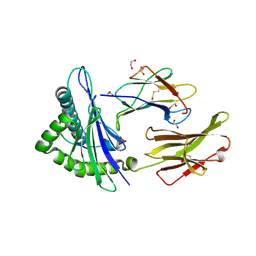 | | Human Leukocyte Antigen A02 presenting ILGKFLHWL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Lloyd, A, Crowther, M, Sewell, A.K. | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Mechanism Underpinning Cross-reactivity of a CD8+ T-cell Clone That Recognizes a Peptide Derived from Human Telomerase Reverse Transcriptase.
J. Biol. Chem., 292, 2017
|
|
6VS7
 
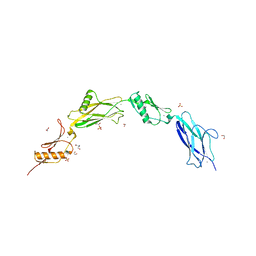 | | Sialic acid binding region of Streptococcus Sanguinis SK1 adhesin | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adhesin, ... | | Authors: | Stubbs, H.E, Iverson, T.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tandem sialoglycan-binding modules in a Streptococcus sanguinis serine-rich repeat adhesin create target dependent avidity effects.
J.Biol.Chem., 295, 2020
|
|
7S3W
 
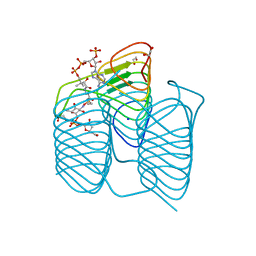 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S44
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
6MVD
 
 | | Crystal structure of Lecithin:cholesterol acyltransferase (LCAT) in complex with isopropyl dodec-11-enylfluorophosphonate (IDFP) and a small molecule activator | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-{4-[(4R)-4-hydroxy-6-oxo-4-(trifluoromethyl)-4,5,6,7-tetrahydro-2H-pyrazolo[3,4-b]pyridin-3-yl]piperidin-1-yl}-4-(trifluoromethyl)pyridine-3-carbonitrile, NICKEL (II) ION, ... | | Authors: | Manthei, K.A, Chang, L, Tesmer, J.J.G. | | Deposit date: | 2018-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis for activation of lecithin:cholesterol acyltransferase by a compound that increases HDL cholesterol.
Elife, 7, 2018
|
|
7P85
 
 | |
6PO4
 
 | | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII. | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII.
To Be Published
|
|
6Y5N
 
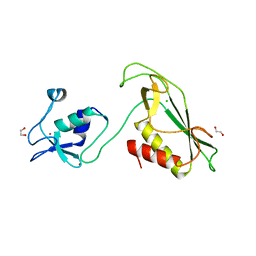 | | RING-DTC domain of Deltex1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase DTX1, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases.
Sci Adv, 6, 2020
|
|
6Y66
 
 | | Structure of Goose Hemorrhagic Polyomavirus VP1 in complex with 2-O-Methyl-5-N-acetyl-alpha-D-neuraminic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, Capsid protein VP1, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
8OWJ
 
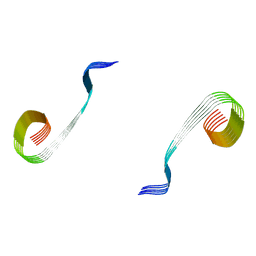 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8G7B
 
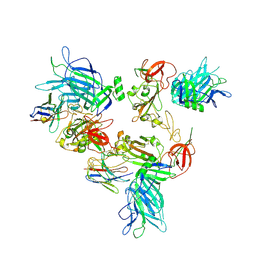 | |
8OVM
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
6EH7
 
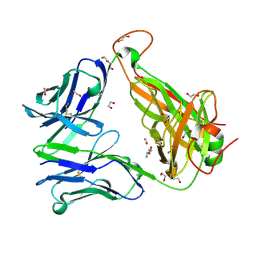 | |
8OWE
 
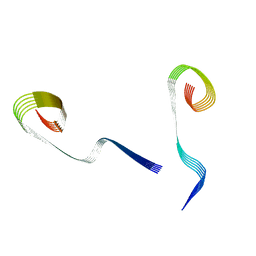 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
