6XAM
 
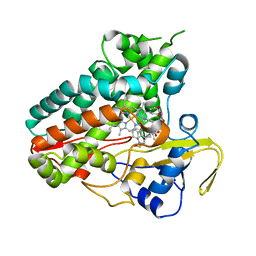 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-homoalanine) | | Descriptor: | (3S,6S)-3-ethyl-6-[(1H-indol-3-yl)methyl]piperazine-2,5-dione, 1,2-ETHANEDIOL, NzeB, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
8OXW
 
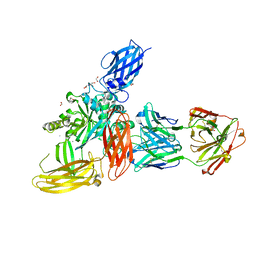 | |
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8OXX
 
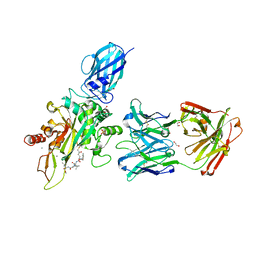 | |
6XCT
 
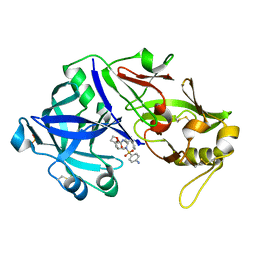 | | Porcine pepsin in complex with amprenavir | | Descriptor: | Pepsin A, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Porcine pepsin in complex with amprenavir
To Be Published
|
|
7RJQ
 
 | | Crystal structure of human Bromodomain containing protein 4 (BRD4) in complex with ILF3 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
To Be Published
|
|
6XD2
 
 | | Porcine pepsin in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Pepsin A | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Porcine pepsin in complex with darunavir
To Be Published
|
|
7RJM
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with ILF3 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, Interleukin enhancer-binding factor 3 | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
8OXV
 
 | |
7P0H
 
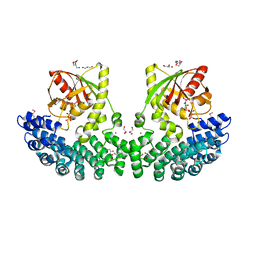 | | Crystal structure of Helicobacter pylori ComF fused to an artificial alphaREP crystallization helper(named B2) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, Helicobacter pylori ComF fused to an artificial alphaREP crystallization helper (named B2), ... | | Authors: | Celma, L, Walbott, H, Legrand, P, Quevillon-Cheruel, S. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | ComFC mediates transport and handling of single-stranded DNA during natural transformation.
Nat Commun, 13, 2022
|
|
8U4T
 
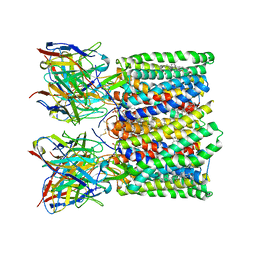 | | Structure of tetrameric CXCR4 in complex with REGN7663 Fab | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, C-X-C chemokine receptor type 4, CHOLESTEROL, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6MME
 
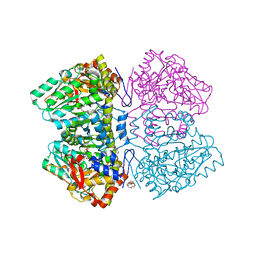 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from S-ethyl-L-cysteine | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
5J63
 
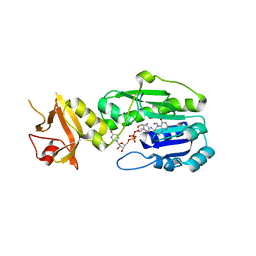 | | Crystal Structure of the N-terminal N-formyltransferase Domain (residues 1-306) of Escherichia coli Arna in Complex with UDP-Ara4N and Folinic Acid | | Descriptor: | (2R,3R,4S,5S)-5-amino-3,4-dihydroxytetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, (4aS,6S)-2-amino-6-{(E)-[(4-methylphenyl)imino]methyl}-4-oxo-4,6,7,8-tetrahydropteridine-5(4aH)-carbaldehyde, Bifunctional polymyxin resistance protein ArnA | | Authors: | Thoden, J.B, Genthe, N.A, Holden, H.M. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Escherichia coli ArnA N-formyltransferase domain in complex with N(5) -formyltetrahydrofolate and UDP-Ara4N.
Protein Sci., 25, 2016
|
|
7OU6
 
 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | Protein O-GlcNAcase, ~{N}-[(3~{Z},6~{S},7~{R},8~{R},8~{a}~{S})-7,8-bis(oxidanyl)-3-(phenylmethyl)imino-1,5,6,7,8,8~{a}-hexahydro-[1,3]thiazolo[3,4-a]pyridin-6-yl]ethanamide | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
6DQ3
 
 | | Streptococcus pyogenes deacetylase PplD in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides
Nat Commun, 13, 2022
|
|
6MY4
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98M,HC-G99M,LC-S30bR] | | Descriptor: | 1,2-ETHANEDIOL, anti-VEGF-A Fab fragment bH1 heavy chain, anti-VEGF-A Fab fragment bH1 light chain | | Authors: | Shi, R, Picard, M.-E, Manenda, M. | | Deposit date: | 2018-11-01 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
7OPA
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzylthiopurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzylthio-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
8TR4
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20404 | | Descriptor: | 4-(2-{(4M)-4-[(6M)-6-(2,5-dimethoxyphenyl)pyridin-3-yl]-1H-1,2,3-triazol-1-yl}ethyl)-N,N-dimethylbenzamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8U4S
 
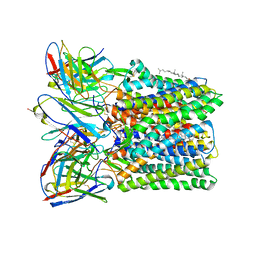 | | Structure of trimeric CXCR4 in complex with REGN7663 Fab | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, C-X-C chemokine receptor type 4, CHOLESTEROL, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5MLM
 
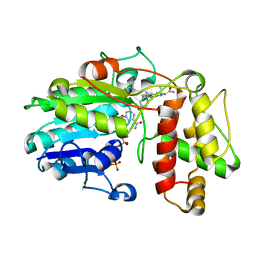 | | Plantago Major multifunctional oxidoreductase V150M mutant in complex with progesterone and NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, Progesterone 5-beta-reductase | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.563 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
8TQD
 
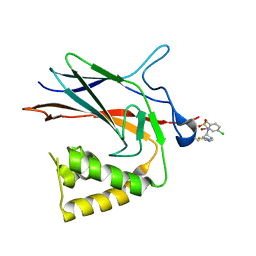 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
8TH6
 
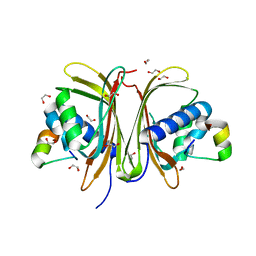 | | Crystal Structure of the G3BP1 NTF2-like domain bound to USP10 peptide | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein-binding protein 1, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Hughes, M.P, Taylor, J.P, Yang, Z. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity.
Cell Rep, 43, 2024
|
|
6VO4
 
 | | Crystal Structure Analysis of BFL1 | | Descriptor: | Bcl-2-related protein A1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2020-01-29 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Identification of a Covalent Molecular Inhibitor of Anti-apoptotic BFL-1 by Disulfide Tethering.
Cell Chem Biol, 27, 2020
|
|
6GI2
 
 | |
8G1H
 
 | | Ancestral protein AncTh of Phosphomethylpirimidine kinases family | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phosphomethylpyrimidine Kinase | | Authors: | Perez, M, Munoz, S, Cea, P, Castro-Fernandez, V. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Deciphering Structural Traits for Thermal and Kinetic Stability across Protein Family Evolution through Ancestral Sequence Reconstruction.
Mol.Biol.Evol., 41, 2024
|
|
