8BS8
 
 | | Bovine naive ultralong antibody AbD08 collected at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heavy chain, Light chain | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The impact of exchanging the light and heavy chains on the structures of bovine ultralong antibodies.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
7AJ9
 
 | |
8J0F
 
 | | GK tetramer with adjacent hooks at reaction state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase B, GAMMA-GLUTAMYL PHOSPHATE, ... | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dynamic Arabidopsis P5CS filament facilitates substrate channelling.
Nat.Plants, 10, 2024
|
|
8S6V
 
 | | Crystal structure of Fab-2D9 chimera complexed to a bis-Tn glycopeptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2D9 (VL-CL), ... | | Authors: | Hurtado-Guerrero, R, Macias-Leon, J, Gonzalez-Ramirez, A. | | Deposit date: | 2024-02-28 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recognizing Tn and STn Epitopes in Protein Context: Structural Advances in Phage Display Libraries for Tumor-Specific Antibody Discovery
Nat.Chem.Biol., 2025
|
|
8UQ7
 
 | | Alzheimer's disease PHF complexed with PET ligand MK-6240 | | Descriptor: | 6-fluoro-3-(1H-pyrrolo[2,3-c]pyridin-1-yl)isoquinolin-5-amine, Microtubule-associated protein tau | | Authors: | Kunach, P, Shahmoradian, S.H, Diamond, M.I, Rosa-Neto, P. | | Deposit date: | 2023-10-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Cryo-EM structure of Alzheimer's disease tau filaments with PET ligand MK-6240.
Nat Commun, 15, 2024
|
|
7Q2I
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Tetrahydrofurfurylamine | | Descriptor: | 1-[(2R)-oxolan-2-yl]methanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7URP
 
 | |
6WIF
 
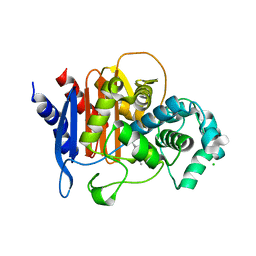 | | Class C beta-lactamase from Acinetobacter baumannii in complex with 4-(Ethyl(methyl)carbamoyl)phenyl boronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | class C beta-lactamase from Acinetobacter baumannii in complex with 4-(Ethyl(methyl)carbamoyl)phenyl boronic acid
To Be Published
|
|
6WDV
 
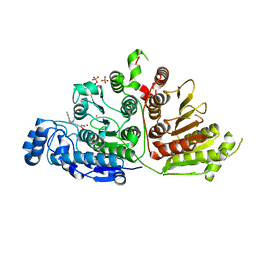 | |
6WLY
 
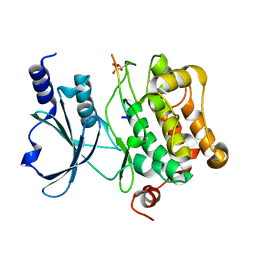 | |
7Q17
 
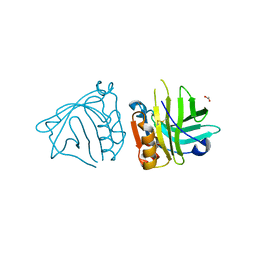 | | Beta-lactoglobulin mutant FAW (I56F/L39A/M107W), unliganded form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin | | Authors: | Loch, J.I, Barciszewski, J, Lewinski, K. | | Deposit date: | 2021-10-18 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
7OPB
 
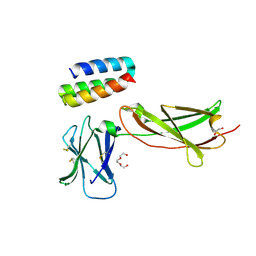 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
6NFN
 
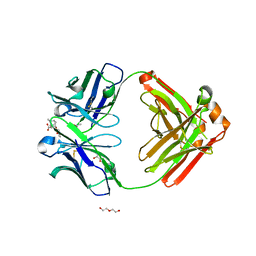 | | Fab fragment of anti-cocaine antibody h2E2 bound to benzoylecgonine | | Descriptor: | 3-(BENZOYLOXY)-8-METHYL-8-AZABICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pokkuluri, P.R, Tan, K. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3D2V
 
 | |
7SWH
 
 | |
6WQ7
 
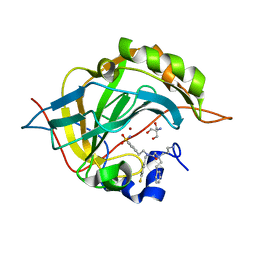 | | Carbonic Anhydrase II Complexed with 2-((3-Aminopropyl)(phenethyl)amino)-N-(4-fluorobenzyl)-N-(4-sulfamoylphenethyl)acetamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, N~2~-(3-aminopropyl)-N-[(4-fluorophenyl)methyl]-N~2~-(2-phenylethyl)-N-[2-(4-sulfamoylphenyl)ethyl]glycinamide, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
6WW1
 
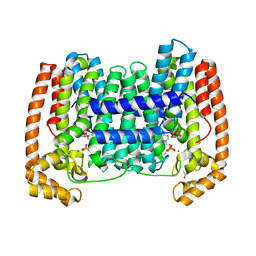 | | Crystal structure of the LmFPPS mutant E97Y | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Gabelli, S.B. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
6WNG
 
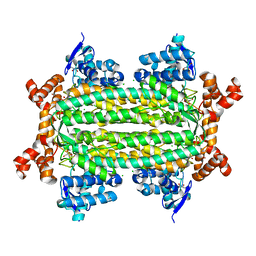 | |
9BGP
 
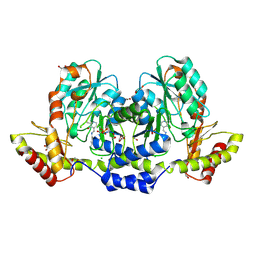 | | X-ray structure of the aminotransferase from Vibrio vulnificus responsible for the biosynthesis of 2,3-diacetamido-4-amino-2,3,4-trideoxy-arabinose in the presence of its internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, aminotransferase | | Authors: | Fait, D.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical Investigation of an Aminotransferase Required for the Production of 2,3,4-triacetamido-2,3,4-trideoxy-L-arabinose
To Be Published
|
|
7BRN
 
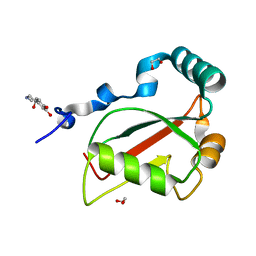 | | Crystal structure of Atg40 AIM fused to Atg8 | | Descriptor: | 1,2-ETHANEDIOL, Autophagy-related protein 40,Autophagy-related protein 8, L-EPINEPHRINE | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7QGJ
 
 | | Apo structure of BIR2 Domain of BIRC2 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Kraemer, A, Farges, F, Schwalm, M.P, Saxena, K, Preuss, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Apo structure BIR2 Domain of BIRC2
To Be Published
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8K3U
 
 | | S. cerevisiae Chs1 in complex with UDP and GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin synthase 1, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8K3R
 
 | | S. cerevisiae Chs1 in apo state incubated with GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
