5OY0
 
 | | Structure of synechocystis photosystem I trimer at 2.5A resolution | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nelson, N, Malavath, T, Caspy, I. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis.
Biochim. Biophys. Acta, 1859, 2018
|
|
8T2F
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody N289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N289 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8T2E
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FP3 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8TPJ
 
 | | Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Orange carotenoid-binding protein, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-04 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
6DID
 
 | | HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Turner, H.L, Cottrell, C.A, Oyen, D, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-05-23 | | Release date: | 2018-09-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
1BG9
 
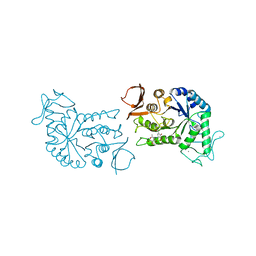 | | BARLEY ALPHA-AMYLASE WITH SUBSTRATE ANALOGUE ACARBOSE | | Descriptor: | 1,4-ALPHA-D-GLUCAN GLUCANOHYDROLASE, 4,6-dideoxy-4-{[(1S,4S,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Kadziola, A, Haser, R. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of a barley alpha-amylase-inhibitor complex: implications for starch binding and catalysis.
J.Mol.Biol., 278, 1998
|
|
8TU6
 
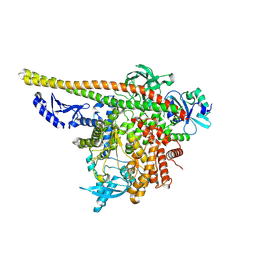 | | CryoEM structure of PI3Kalpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Valverde, R, Shi, H, Holliday, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8U66
 
 | | Firmicutes Rubisco | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Rubisco | | Authors: | Kaeser, B.P, Liu, A.K, Shih, P.M. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Deep-branching evolutionary intermediates reveal structural origins of form I rubisco.
Curr.Biol., 33, 2023
|
|
8TSA
 
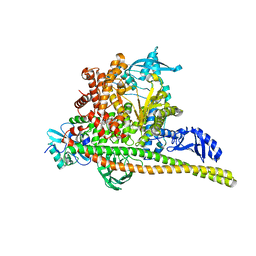 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 2 | | Descriptor: | 5-(3-bromo-5-fluorobenzamido)-N-methyl-6-(2-methylanilino)pyridine-3-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J, Valverde, R. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TSB
 
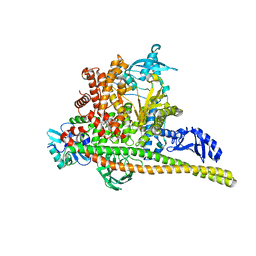 | | Human PI3K p85alpha/p110alpha bound to compound 2 | | Descriptor: | 5-(3-bromo-5-fluorobenzamido)-N-methyl-6-(2-methylanilino)pyridine-3-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TS8
 
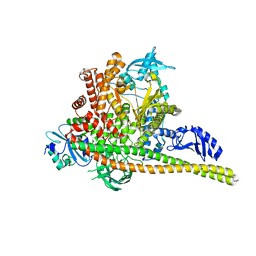 | | p85alpha/p110alpha heterodimer H1047R mutant | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TSD
 
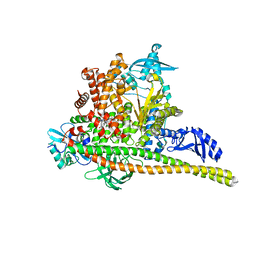 | | Human PI3K p85alpha/p110alpha bound to RLY-2608 | | Descriptor: | N-{(3R,6M)-3-(2-chloro-5-fluorophenyl)-6-[(4S)-5-cyano[1,2,4]triazolo[1,5-a]pyridin-6-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-3-fluoro-5-(trifluoromethyl)benzamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TS7
 
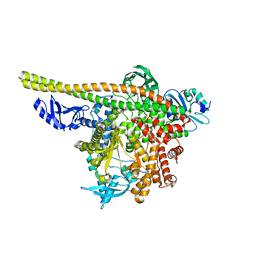 | | Human PI3K p85alpha/p110alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TS9
 
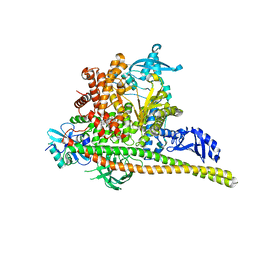 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 1 | | Descriptor: | 5-[3-fluoro-5-(trifluoromethyl)benzamido]-N-methyl-6-(2-methylanilino)pyridine-3-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TSC
 
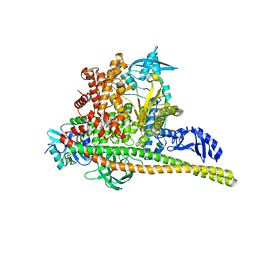 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 3 | | Descriptor: | (1S)-7-[3-fluoro-5-(trifluoromethyl)benzamido]-N-methyl-1-(2-methylphenyl)-3-oxo-2,3-dihydro-1H-isoindole-5-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TO5
 
 | | Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (1.87 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TRO
 
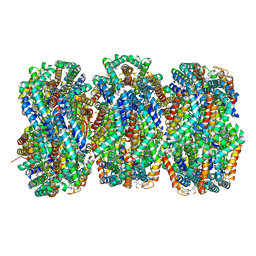 | | Rod from high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TO2
 
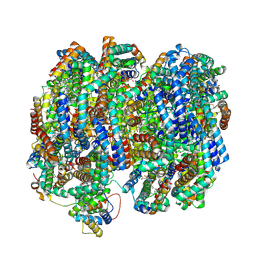 | | Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TWG
 
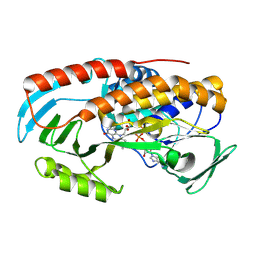 | |
1BOM
 
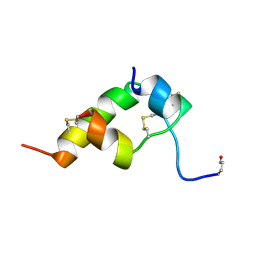 | | THREE-DIMENSIONAL STRUCTURE OF BOMBYXIN-II, AN INSULIN-RELATED BRAIN-SECRETORY PEPTIDE OF THE SILKMOTH BOMBYX MORI: COMPARISON WITH INSULIN AND RELAXIN | | Descriptor: | BOMBYXIN-II,BOMBYXIN A-2, BOMBYXIN-II,BOMBYXIN A-6 | | Authors: | Nagata, K, Hatanaka, H, Kohda, D, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-11-01 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of bombyxin-II an insulin-like peptide of the silkmoth Bombyx mori: structural comparison with insulin and relaxin.
J.Mol.Biol., 253, 1995
|
|
4BV2
 
 | | CRYSTAL STRUCTURE OF SIR2 IN COMPLEX WITH THE INHIBITOR EX-527, 2'-O-ACETYL-ADP-RIBOSE AND DEACETYLATED P53-PEPTIDE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, CELLULAR TUMOR ANTIGEN P53, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
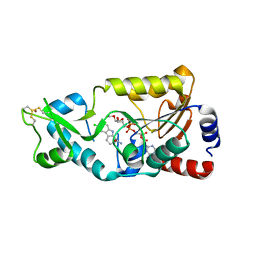
6E5X
 
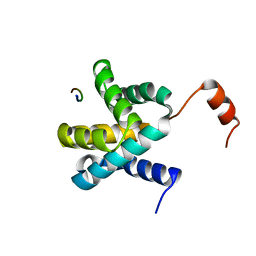 | | Crystal structure of Ebola virus VP30 C-terminus/RBBP6 peptide complex | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase RBBP6, Minor nucleoprotein VP30 | | Authors: | Liu, D, Small, G.I, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-07-23 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication.
Cell, 175, 2018
|
|
1D2C
 
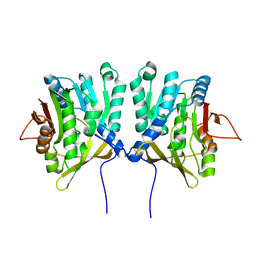 | | METHYLTRANSFERASE | | Descriptor: | PROTEIN (GLYCINE N-METHYLTRANSFERASE) | | Authors: | Huang, Y, Takusagawa, F. | | Deposit date: | 1999-09-23 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
6EFG
 
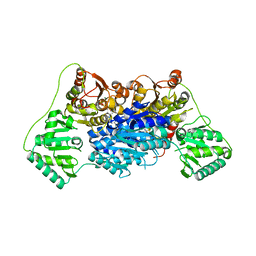 | | Pyruvate decarboxylase from Kluyveromyces lactis | | Descriptor: | MAGNESIUM ION, Pyruvate decarboxylase, THIAMINE DIPHOSPHATE | | Authors: | Kutter, S, Konig, S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structures of pyruvate decarboxylase from Kluyveromyces lactis in the absence of ligands and in the presence of the substrate surrogate pyruvamide
To be Published
|
|
6ES6
 
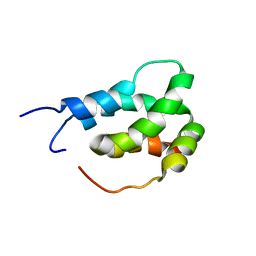 | |
