5F35
 
 | | Structure of quinolinate synthase in complex with citrate | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
5F3D
 
 | | Structure of quinolinate synthase in complex with reaction intermediate W | | Descriptor: | 2-IMINO,3-CARBOXY,5-OXO,6-HYDROXY HEXANOIC ACID, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
2WPW
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (without AcCoA) | | Descriptor: | ACETYL COENZYME *A, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
2WL6
 
 | |
5F33
 
 | | Structure of quinolinate synthase in complex with phosphoglycolohydroxamate | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHOGLYCOLOHYDROXAMIC ACID, Quinolinate synthase A, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
3IQI
 
 | |
6TDH
 
 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 1 | | Descriptor: | 3-(2-chlorophenyl)-4~{H}-1,2,4-triazole, ACETYL COENZYME *A, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
6PUX
 
 | |
5J7W
 
 | |
3O6C
 
 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
4E7B
 
 | |
5ZZU
 
 | |
4E8O
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ih from Acinetobacter baumannii | | Descriptor: | Aac(6')-Ih protein, CHLORIDE ION | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
6A2N
 
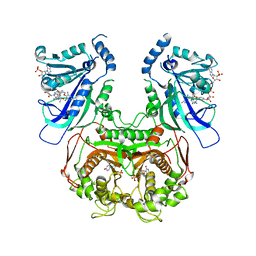 | | Crystal structure of quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum DHFR-TS complexed with BT2, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)-6-ethylpyrimidine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Tarnchampoo, B, Yuthavong, Y. | | Deposit date: | 2018-06-12 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
3ID0
 
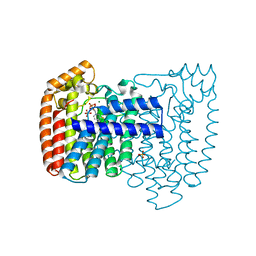 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with 3-Fluoro-1-(2-hydroxy-2,2-bisphosphono-ethyl)pyridinium | | Descriptor: | 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-19 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
5IX3
 
 | |
7XAY
 
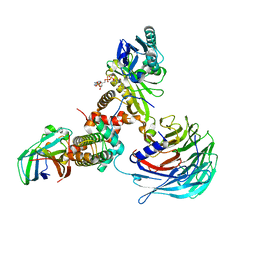 | | Crystal structure of Hat1-Hat2-Asf1-H3-H4 | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yue, Y, Yang, W.S, Xu, R.M. | | Deposit date: | 2022-03-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Topography of histone H3-H4 interaction with the Hat1-Hat2 acetyltransferase complex.
Genes Dev., 36, 2022
|
|
7L4L
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C8-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, SODIUM ION, ... | | Authors: | Mindrebo, J.T, Chen, A, Kim, W.E, Burkart, M.D, Noel, J.P. | | Deposit date: | 2020-12-19 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Mechanistic Analyses of the Gating Mechanism of Elongating Ketosynthases
Acs Catalysis, 11, 2021
|
|
2V07
 
 | | Structure of the Arabidopsis thaliana cytochrome c6A V52Q variant | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Worrall, J.A.R, Schlarb-Ridley, B.G, Reda, T, Marcaida, M.J, Moorlen, R.J, Wastl, J, Hirst, J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of heme redox potential in the cytochrome c6 family.
J. Am. Chem. Soc., 129, 2007
|
|
5F0V
 
 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-11-28 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5F46
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample, apoenzyme form | | Descriptor: | CHLORIDE ION, aminoglycoside acetyltransferase meta-AAC0020 | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
3I8P
 
 | |
3NVS
 
 | | 1.02 Angstrom resolution crystal structure of 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae in complex with shikimate-3-phosphate (partially photolyzed) and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, G, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.021 Å) | | Cite: | 1.02 Angstrom Resolution Crystal Structure of 3-Phosphoshikimate 1-Carboxyvinyltransferase from Vibrio cholerae in complex with Shikimate-3-Phosphate (Partially Photolyzed) and Glyphosate
TO BE PUBLISHED
|
|
5JKJ
 
 | | Crystal structure of esterase E22 L374D mutant | | Descriptor: | Esterase E22 | | Authors: | Zhang, Y, Wang, P, Yao, Q. | | Deposit date: | 2016-04-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for substrate recognition and catalysis of a novel esterase E22 with a homoserine transacetylase-like fold
To Be Published
|
|
5F48
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, MAGNESIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
