2M9I
 
 | | NMR solution structure of Pin1 WW domain variant 6-1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-10 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
6J3Y
 
 | | Structure of C2S2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
4PCO
 
 | |
2MU4
 
 | | Structure of F. tularensis Virulence Determinant | | Descriptor: | flpp3Sol_2 | | Authors: | Zook, J.J.D.Z, Mo, G.G.M, Craciunescu, F.F.C, Sisco, N.N.S, Hansen, D.D.H, Baravati, B.B.B, Van Horn, W.W.V.H, Cherry, B.B.C, Fromme, P.P.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Francisella tularensis Virulence Determinant Reveals Structural Homology to Bet v1 Allergen Proteins.
Structure, 23, 2015
|
|
6A2W
 
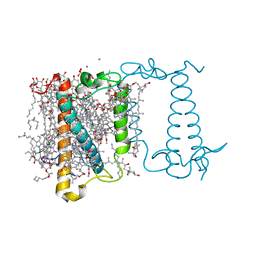 | | Crystal structure of fucoxanthin chlorophyll a/c complex from Phaeodactylum tricornutum | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W, Yu, L.J, Kuang, T.Y, Shen, J.R. | | Deposit date: | 2018-06-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for blue-green light harvesting and energy dissipation in diatoms.
Science, 363, 2019
|
|
2NCN
 
 | |
7XXH
 
 | | Cryo-EM structure of the purinergic receptor P2Y1R in complex with 2MeSADP and G11 | | Descriptor: | 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), Guanine nucleotide-binding protein G(11) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tan, Q, Li, B, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R.
Protein Cell, 14, 2023
|
|
4NVB
 
 | |
4NVM
 
 | |
4NVG
 
 | |
2CGI
 
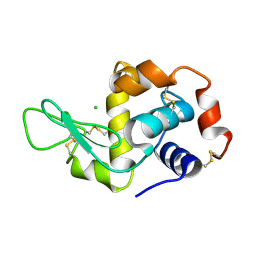 | | Siras structure of tetragonal lysozyme using derivative data collected at the high energy remote Holmium Kedge | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Jakoncic, J, Di Michiel, M, Zhong, Z, Honkimaki, V, Jouanneau, Y, Stojanoff, V. | | Deposit date: | 2006-03-07 | | Release date: | 2006-11-13 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Anomalous Diffraction at Ultra-High Energy for Protein Crystallography.
J.Appl.Crystallogr., 39, 2006
|
|
4NVJ
 
 | |
4NVF
 
 | |
4NVE
 
 | |
4NVO
 
 | |
4NVN
 
 | | Predicting protein conformational response in prospective ligand discovery | | Descriptor: | 2,3-dihydrobenzo[h][1,6]naphthyridin-4(1H)-one, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery.
Nat Chem, 6, 2014
|
|
6J40
 
 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
4NVC
 
 | |
8P25
 
 | | Solution structure of a chimeric U2AF2 RRM2 / FUBP1 N-Box | | Descriptor: | Splicing factor U2AF 65 kDa subunit,Far upstream element-binding protein 1 | | Authors: | Hipp, C, Sattler, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | FUBP1 is a general splicing factor facilitating 3' splice site recognition and splicing of long introns.
Mol.Cell, 83, 2023
|
|
4NVI
 
 | |
4NVD
 
 | |
4NVA
 
 | |
4NVL
 
 | |
4NVK
 
 | | Predicting protein conformational response in prospective ligand discovery. | | Descriptor: | Cytochrome c peroxidase, N~2~,N~2~-diethylquinazoline-2,4-diamine, PHOSPHATE ION, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery.
Nat Chem, 6, 2014
|
|
4NVH
 
 | |
