8X21
 
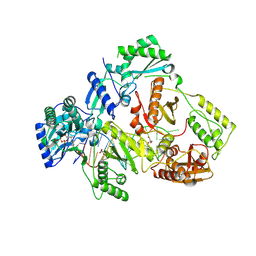 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:ETV-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
6RWM
 
 | | SIVrcm intasome in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RFS
 
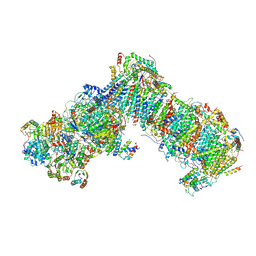 | | Cryo-EM structure of a respiratory complex I mutant lacking NDUFS4 | | Descriptor: | Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), Acyl carrier protein ACPM2 of NADH:Ubiquinone Oxidoreductase (Complex I), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2019-04-16 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease.
Sci Adv, 5, 2019
|
|
5WJU
 
 | | Cryo-EM structure of B. subtilis flagellar filaments A39V, N133H | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
7WFE
 
 | | Right PSI in the cyclic electron transfer supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
4Z3S
 
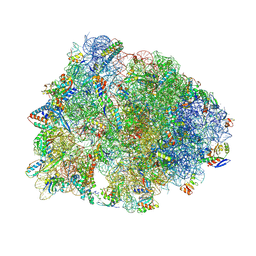 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic A201A, mRNA and three tRNAs in the A, P and E sites at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 3'-deoxy-3'-{[(2E)-3-(4-{[(4Z)-6-O-(6-deoxy-3,4-di-O-methyl-alpha-D-mannopyranosyl)-5-O-methyl-alpha-D-threo-hex-4-enofuranosyl]oxy}phenyl)-2-methylprop-2-enoyl]amino}-N,N-dimethyladenosine, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-03-31 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
6RXZ
 
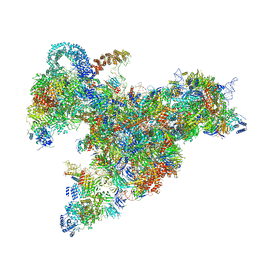 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state b | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S11-like protein, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
5WOB
 
 | | Crystal Structure Analysis of Fab1-Bound Human Insulin Degrading Enzyme (IDE) in Complex with Insulin | | Descriptor: | IDE-bound Fab heavy chain, IDE-bound Fab light chain, Insulin, ... | | Authors: | McCord, L.A, Liang, W.G, Farcasanu, M, Wang, A.G, Koide, S, Tang, W.J. | | Deposit date: | 2017-08-01 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
4YY8
 
 | | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum | | Descriptor: | GLYCEROL, Kelch protein, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, El Bakkouri, M, Senisterra, G, Osman, K.T, Lovato, D.V, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum.
to be published
|
|
7WPF
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with three JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
6KF7
 
 | | Microbial Hormone-sensitive lipase E53 mutant S285G | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Xiaochen, Y, Zhengyang, L, Xuewei, X, Jixi, L. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Microbial Hormone-sensitive lipase E53 mutant S285G
To Be Published
|
|
4YZD
 
 | | Crystal Structure of human phosphorylated IRE1alpha in complex with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
6S38
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
7WFD
 
 | | Left PSI in the cyclic electron transport supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pan, X, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
6KI8
 
 | | Pyrophosphatase mutant K149R from Acinetobacter baumannii | | Descriptor: | DIPHOSPHATE, Inorganic pyrophosphatase, MAGNESIUM ION | | Authors: | Su, J. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
4Z40
 
 | | Active site complex BamBC of Benzoyl Coenzyme A reductase as isolated | | Descriptor: | Benzoyl-CoA reductase, putative, IRON/SULFUR CLUSTER, ... | | Authors: | Weinert, T, Kung, J.W, Weidenweber, S, Huwiler, S.G, Boll, M, Ermler, U. | | Deposit date: | 2015-04-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
7WPE
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with two JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
5UYK
 
 | | 70S ribosome bound with cognate ternary complex not base-paired to A site codon (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
4ZGE
 
 | | Double Mutant H80W/H81W of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-22 | | Release date: | 2015-07-01 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
8X20
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:E-CFCP-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), E-CFCP-triphosphate, GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
5V5X
 
 | | Protocadherin gammaB7 EC3-6 cis-dimer structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Protocadherin cis-dimer architecture and recognition unit diversity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6S47
 
 | | Saccharomyces cerevisiae 80S ribosome bound with ABCF protein New1 | | Descriptor: | 18S rRNA (1707-MER), 28S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Kasari, V, Pochopien, A.A, Margus, T, Murina, V, Turnbull, K, Zhou, Y, Nissan, T, Graf, M, Novacek, J, Atkinson, G.C, Johansson, M.J.O, Wilson, D.N, Hauryliuk, V. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A role for the Saccharomyces cerevisiae ABCF protein New1 in translation termination/recycling.
Nucleic Acids Res., 47, 2019
|
|
6RKC
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, Methionine adenosyltransferase, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6JT7
 
 | | Crystal structure of 452-453_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
5UJG
 
 | | ovGRN12-35_3s | | Descriptor: | Granulin | | Authors: | Bansal, P, Smout, M, Wilson, D, Caceres, C.C, Dastpeyman, M, Sotillo, J, Seifert, J, Brindley, P, Loukas, A, Daly, N. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Development of a Potent Wound Healing Agent Based on the Liver Fluke Granulin Structural Fold.
J. Med. Chem., 60, 2017
|
|
