3CT2
 
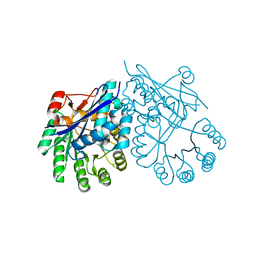 | | Crystal structure of muconate cycloisomerase from Pseudomonas fluorescens | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes.
Biochemistry, 48, 2009
|
|
2QY0
 
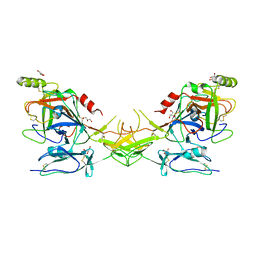 | | Active dimeric structure of the catalytic domain of C1r reveals enzyme-product like contacts | | Descriptor: | Complement C1r subcomponent, GLYCEROL | | Authors: | Kardos, J, Harmat, V, Pallo, A, Barabas, O, Szilagyi, K, Graf, L, Naray-Szabo, G, Goto, Y, Zavodszky, P, Gal, P. | | Deposit date: | 2007-08-13 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r.
Mol.Immunol., 45, 2008
|
|
1S7W
 
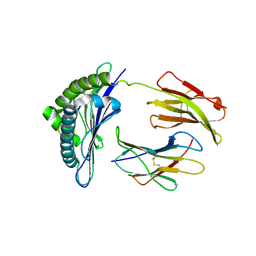 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1RYY
 
 | |
2QTJ
 
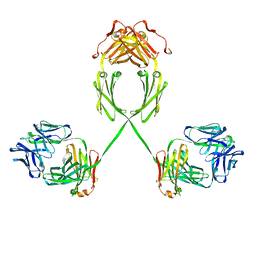 | | Solution structure of human dimeric immunoglobulin A | | Descriptor: | Ig alpha-1 chain C region, Kappa light chain IgA1 | | Authors: | Bonner, A, Furtado, P.B, Almogren, A, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Implications of the near-planar solution structure of human myeloma dimeric IgA1 for mucosal immunity and IgA nephropathy
J.Immunol., 180, 2008
|
|
3CW9
 
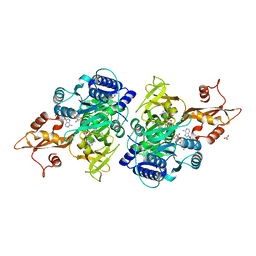 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase in the Thioester-forming Conformation, bound to 4-chlorophenacyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-Chlorophenacyl-coenzyme A, 4-chlorobenzoyl CoA ligase, ... | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
2QBY
 
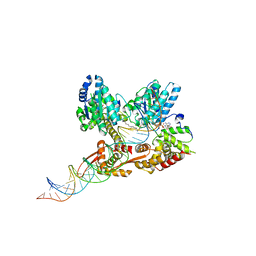 | | Crystal structure of a heterodimer of Cdc6/Orc1 initiators bound to origin DNA (from S. solfataricus) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 6 homolog 1, Cell division control protein 6 homolog 3, ... | | Authors: | Cunningham Dueber, E.L, Corn, J.E, Bell, S.D, Berger, J.M. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex.
Science, 317, 2007
|
|
3D18
 
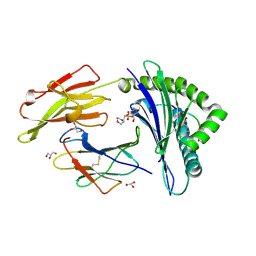 | | Crystal structure of HLA-B*2709 complexed with a variant of the latent membrane protein 2 peptide (LMP2(L)) of epstein-barr virus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, ... | | Authors: | Beltrami, A, Gabdulkhakov, A, Rossmann, M, Ziegler, A, Uchanska-Ziegler, B, Saenger, W. | | Deposit date: | 2008-05-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of HLA-B*2709 complexed with a variant of the latent membrane protein 2 peptide (LMP2(L)) of epstein-barr virus
To be Published
|
|
2QBS
 
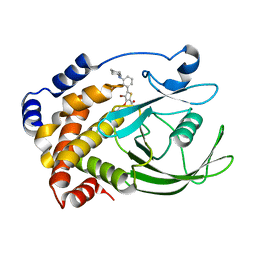 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)-5-[3-(CYCLOHEXYLAMINO)PHENYL]THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
3CXZ
 
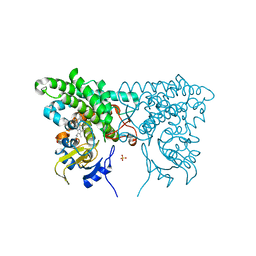 | |
2QDJ
 
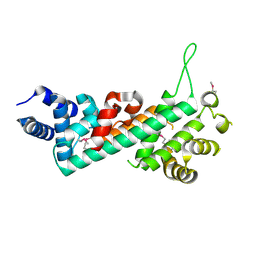 | | Crystal structure of the Retinoblastoma protein N-domain provides insight into tumor suppression, ligand interaction and holoprotein architecture | | Descriptor: | Retinoblastoma-associated protein | | Authors: | Hassler, M, Mittnacht, S, Pearl, L.H. | | Deposit date: | 2007-06-21 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retinoblastoma protein N domain provides insight into tumor suppression, ligand interaction, and holoprotein architecture.
Mol.Cell, 28, 2007
|
|
3D2E
 
 | | Crystal structure of a complex of Sse1p and Hsp70, Selenomethionine-labeled crystals | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Heat shock 70 kDa protein 1, ... | | Authors: | Polier, S, Bracher, A. | | Deposit date: | 2008-05-08 | | Release date: | 2008-06-17 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding.
Cell(Cambridge,Mass.), 133, 2008
|
|
1RZF
 
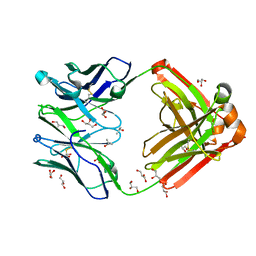 | | Crystal structure of Human anti-HIV-1 GP120-reactive antibody E51 | | Descriptor: | Fab E51 heavy chain, Fab E51 light chain, GLYCEROL, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3D4O
 
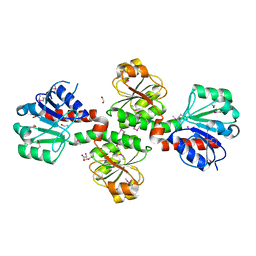 | |
3D59
 
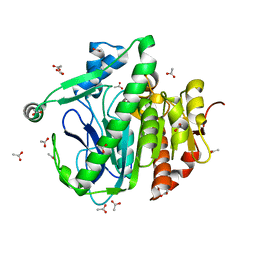 | |
3D1N
 
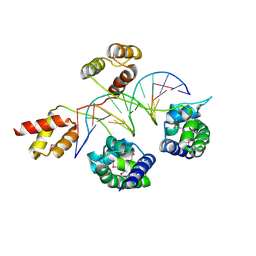 | | Structure of human Brn-5 transcription factor in complex with corticotrophin-releasing hormone gene promoter | | Descriptor: | 5'-D(*DAP*DGP*DCP*DAP*DTP*DAP*DAP*DAP*DTP*DAP*DAP*DTP*DAP*DA)-3', 5'-D(*DTP*DTP*DAP*DTP*DTP*DAP*DTP*DTP*DTP*DAP*DTP*DGP*DCP*DT)-3', POU domain, ... | | Authors: | Pereira, J.H, Ha, S.C, Kim, S.-H. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of human Brn-5 transcription factor in complex with CRH gene promoter.
J.Struct.Biol., 167, 2009
|
|
1SBG
 
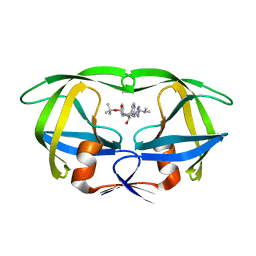 | |
3CF2
 
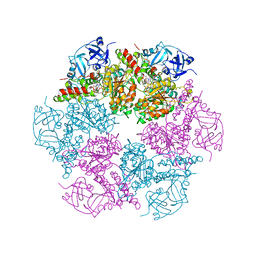 | | Structure of P97/vcp in complex with ADP/AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Davies, J.M, Delabarre, B, Brunger, A.T, Weis, W.I. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.
Structure, 16, 2008
|
|
2QI1
 
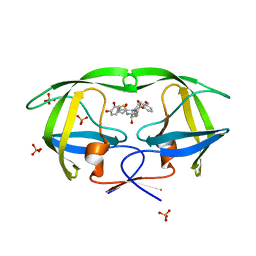 | | Crystal structure of protease inhibitor, MIT-1-KK81 in complex with wild type HIV-1 protease | | Descriptor: | N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{[(3-METHOXYPHENYL)SULFONYL](2-THIENYLMETHYL)AMINO}PROPYL]-3,4-DIHYDROXYBENZAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QMI
 
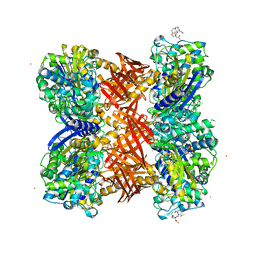 | | Structure of the octameric penicillin-binding protein homologue from Pyrococcus abyssi | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, LUTETIUM (III) ION, Pbp related beta-lactamase | | Authors: | Delfosse, V, Girard, E, Moulinier, L, Schultz, P, Mayer, C. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the archaeal pab87 peptidase reveals a novel self-compartmentalizing protease family
Plos One, 4, 2009
|
|
1RXD
 
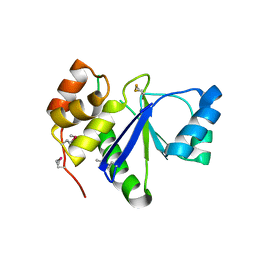 | | Crystal structure of human protein tyrosine phosphatase 4A1 | | Descriptor: | protein tyrosine phosphatase type IVA, member 1; Protein tyrosine phosphatase IVA1 | | Authors: | Sun, J.P, Fedorov, A.A, Almo, S.C, Zhang, Z.Y, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-18 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1RXS
 
 | | E. coli uridine phosphorylase: 2'-deoxyuridine phosphate complex | | Descriptor: | 2'-DEOXYURIDINE, META VANADATE, PHOSPHATE ION, ... | | Authors: | Caradoc-Davies, T.T, Cutfield, S.M, Lamont, I.L, Cutfield, J.F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium
J.Mol.Biol., 337, 2004
|
|
3CIS
 
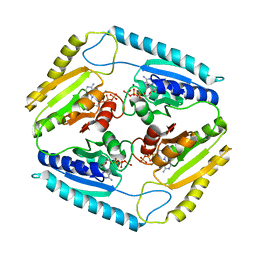 | | The Crystal Structure of Rv2623 from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein | | Authors: | Bilder, P, Drumm, J, Mi, K, Chan, J, Almo, S.C. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of Rv2623 : a Novel, Tandem-Repeat Universal Stress Protein of Mycobacterium tuberculosis
To be Published
|
|
2QQR
 
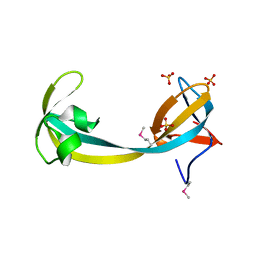 | | JMJD2A hybrid tudor domains | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, SULFATE ION | | Authors: | Lee, J, Botuyan, M.V, Mer, G. | | Deposit date: | 2007-07-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3CKC
 
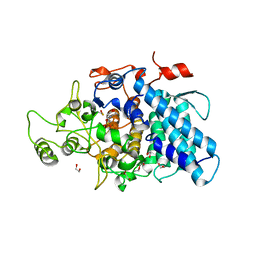 | | B. thetaiotaomicron SusD | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
