2ZDP
 
 | | Crystal structure of IsdI in complex with Cobalt protoporphyrin IX | | Descriptor: | CHLORIDE ION, Heme-degrading monooxygenase isdI, PROTOPORPHYRIN IX CONTAINING CO | | Authors: | Lee, W.C, Reniere, M.L, Skaar, E.P, Murphy, M.E.P. | | Deposit date: | 2007-11-27 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ruffling of Metalloporphyrins Bound to IsdG and IsdI, Two Heme-degrading Enzymes in Staphylococcus aureus
J.Biol.Chem., 283, 2008
|
|
2ZU6
 
 | | crystal structure of the eIF4A-PDCD4 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Cho, Y, Chang, J.H, Sohn, S.Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the eIF4A-PDCD4 complex
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
2P1U
 
 | | Crystal structure of the ligand binding domain of the retinoid X receptor alpha in complex with 3-(2'-ethoxy)-tetrahydronaphtyl cinnamic acid and a fragment of the coactivator TIF-2 | | Descriptor: | (2E)-3-[3-(3-ETHOXY-5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)-4-HYDROXYPHENYL]ACRYLIC ACID, Nuclear receptor coactivator 2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Bourguet, W, Nahoum, V. | | Deposit date: | 2007-03-06 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1QOX
 
 | |
5VEY
 
 | | Solution NMR structure of histone H2A-H2B mono-ubiquitylated at H2A Lys15 in complex with RNF169 (653-708) | | Descriptor: | E3 ubiquitin-protein ligase RNF169, Histone H2B type 1-J,Histone H2A type 1-B/E, Polyubiquitin-B | | Authors: | Hu, Q, Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18.
Mol. Cell, 66, 2017
|
|
3J3T
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J4U
 
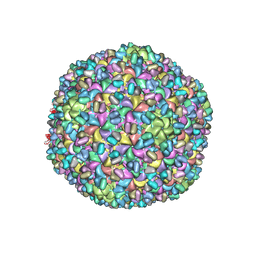 | | A new topology of the HK97-like fold revealed in Bordetella bacteriophage: non-covalent chainmail secured by jellyrolls | | Descriptor: | cementing protein, major capsid protein | | Authors: | Zhang, X, Guo, H, Jin, L, Czornyj, E, Hodes, A, Hui, W.H, Nieh, A.W, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution.
Elife, 2, 2013
|
|
2OWN
 
 | |
1QUB
 
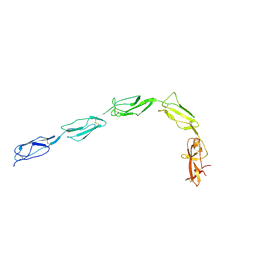 | | CRYSTAL STRUCTURE OF THE GLYCOSYLATED FIVE-DOMAIN HUMAN BETA2-GLYCOPROTEIN I PURIFIED FROM BLOOD PLASMA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (human beta2-Glycoprotein I), ... | | Authors: | Bouma, B, de Groot, P.G, van den Elsen, J.M.H, Ravelli, R.B.G, Schouten, A, Simmelink, M.J.A, Derksen, R.H.W.M, Kroon, J, Gros, P. | | Deposit date: | 1999-07-01 | | Release date: | 1999-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Adhesion mechanism of human beta(2)-glycoprotein I to phospholipids based on its crystal structure.
EMBO J., 18, 1999
|
|
3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
5VHX
 
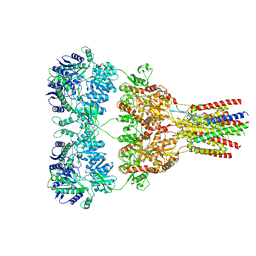 | | GluA2-1xGSG1L bound to ZK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes.
Neuron, 94, 2017
|
|
3J6G
 
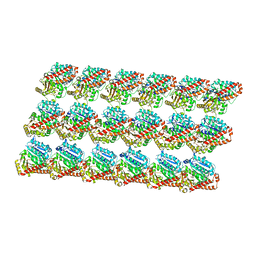 | | Minimized average structure of microtubules stabilized by taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-19 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
1QBU
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R--(1ALPHA,5ALPHA,7BETA)]-3-[(CYCLOPROPHYLMETHYL)HEXAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPIN] METHYL-2-THIAZOLYLBENZAMIDE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
3ASQ
 
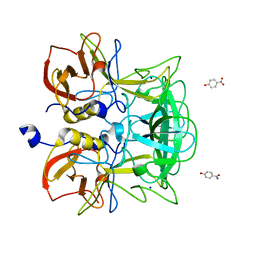 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with H-antigen | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
5VJH
 
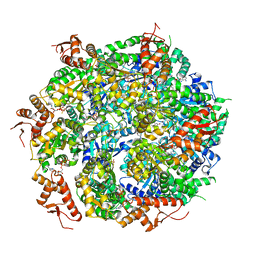 | | Closed State CryoEM Reconstruction of Hsp104:ATPyS and FITC casein | | Descriptor: | FITC casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
3J6Q
 
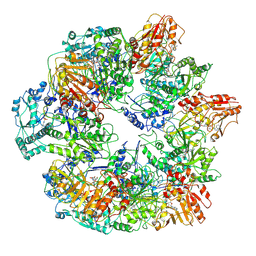 | | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Structural protein VP3 | | Authors: | Zhu, B, Yang, C, Liu, H, Cheng, L, Song, F, Zeng, S, Huang, X, Ji, G, Zhu, P. | | Deposit date: | 2014-03-20 | | Release date: | 2014-10-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus.
J.Mol.Biol., 426, 2014
|
|
5GM7
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina at 1.78 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Ribosome inactivating protein | | Authors: | Singh, B, Singh, P.K, Pandey, S.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-07-13 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina at 1.78 Angstrom resolution
To Be Published
|
|
3J96
 
 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
1QM8
 
 | | Structure of Bacteriorhodopsin at 100 K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Kamiya, N, Kouyama, T. | | Deposit date: | 1999-09-22 | | Release date: | 2000-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Three-Dimensional Crystal of Bacteriorhodopsin Obtained by Successive Fusion of the Vesicular Assemblies.
J.Mol.Biol., 283, 1998
|
|
3JAR
 
 | | Cryo-EM structure of GDP-microtubule co-polymerized with EB3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, R, Nogales, E. | | Deposit date: | 2015-06-19 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins.
Cell(Cambridge,Mass.), 162, 2015
|
|
1NNF
 
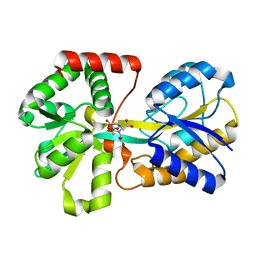 | | Crystal Structure Analysis of Haemophlius Influenzae Ferric-ion Binding Protein H9Q Mutant Form | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Shouldice, S.R, Dougan, D.R, Skene, R.J, Tari, L.W, McRee, D.E, Yu, R.-H, Schryvers, A.B. | | Deposit date: | 2003-01-13 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High Resolution Structure of an Alternate Form of the Ferric ion Binding Protein from Haemophilus influenzae
J.Biol.Chem., 278, 2003
|
|
5GS0
 
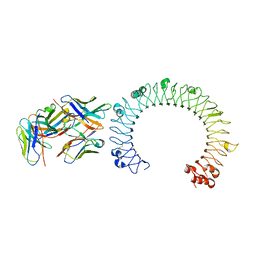 | | Crystal structure of the complex of TLR3 and bi-specific diabody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 3, alpha-D-mannopyranose, ... | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.275 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
2PAN
 
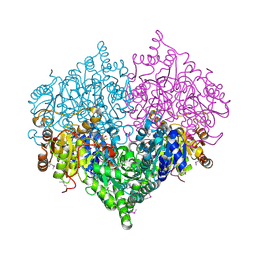 | | Crystal structure of E. coli glyoxylate carboligase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kaplun, A, Chipman, D.M, Barak, Z, Vyazmensky, M, Shaanan, B. | | Deposit date: | 2007-03-27 | | Release date: | 2008-01-01 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glyoxylate carboligase lacks the canonical active site glutamate of thiamine-dependent enzymes.
Nat.Chem.Biol., 4, 2008
|
|
3J8D
 
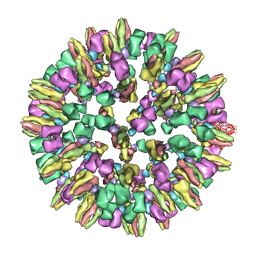 | | Cryoelectron microscopy of dengue-Fab E104 complex at pH 5.5 | | Descriptor: | Envelope protein E, antibody E111 Fab fragment, glycoprotein DIII | | Authors: | Zhang, X.Z, Sheng, J, Austin, S.K, Hoornweg, T, Smit, J.M, Kuhn, R.J, Diamond, M.S, Rossmann, M.G. | | Deposit date: | 2014-10-13 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Structure of Acidic pH Dengue Virus Showing the Fusogenic Glycoprotein Trimers.
J.Virol., 89, 2015
|
|
3J9X
 
 | | A Virus that Infects a Hyperthermophile Encapsidates A-Form DNA | | Descriptor: | DNA, coat protein | | Authors: | DiMaio, F, Yu, X, Rensen, E, Krupovic, M, Prangishvili, D, Egelman, E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A virus that infects a hyperthermophile encapsidates A-form DNA.
Science, 348, 2015
|
|
