3UGB
 
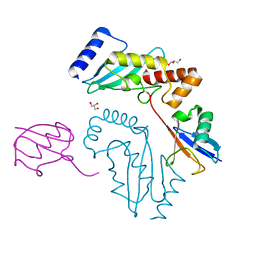 | | UbcH5c~Ubiquitin Conjugate | | Descriptor: | GLYCEROL, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Page, R.C, Pruneda, J.N, Klevit, R.E, Misra, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the conformation and oligomerization of E2~ubiquitin conjugates.
Biochemistry, 51, 2012
|
|
1N5U
 
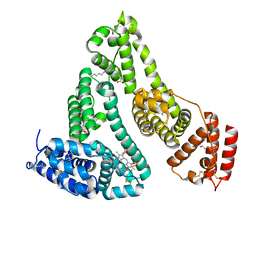 | | X-RAY STUDY OF HUMAN SERUM ALBUMIN COMPLEXED WITH HEME | | Descriptor: | MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SERUM ALBUMIN | | Authors: | Wardell, M, Wang, Z, Ho, J.X, Robert, J, Ruker, F, Ruble, J, Carter, D.C. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Atomic Structure of Human Methemalbumin at 1.9 A
Biochem.Biophys.Res.Commun., 291, 2002
|
|
3UUB
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1S40
 
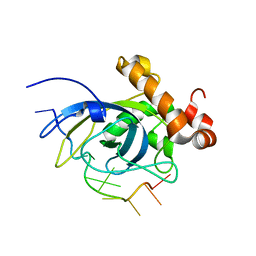 | | SOLUTION STRUCTURE OF THE CDC13 DNA-BINDING DOMAIN COMPLEXED WITH A SINGLE-STRANDED TELOMERIC DNA 11-MER | | Descriptor: | 5'-D(*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G)-3', Cell division control protein 13 | | Authors: | Mitton-Fry, R.M, Anderson, E.M, Theobald, D.L, Glustrom, L.W, Wuttke, D.S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13
J.Mol.Biol., 338, 2004
|
|
3US0
 
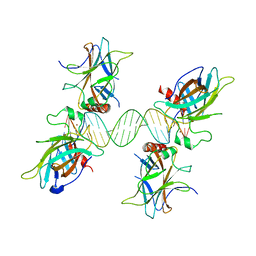 | |
2AVT
 
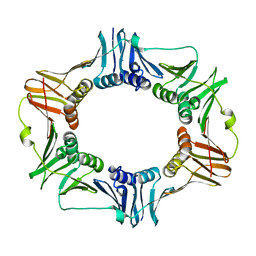 | | Crystal structure of the beta subunit from DNA polymerase of Streptococcus pyogenes | | Descriptor: | DNA polymerase III beta subunit | | Authors: | Argiriadi, M.A, Goedken, E.R, Bruck, I, O'donnell, M, Kuriyan, J. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium.
Bmc Struct.Biol., 6, 2006
|
|
2B1L
 
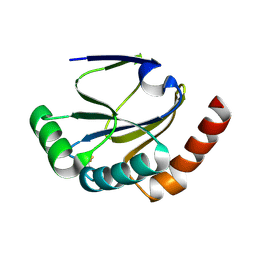 | |
2DRU
 
 | | Crystal structure and binding properties of the CD2 and CD244 (2B4) binding protein, CD48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, chimera of CD48 antigen and T-cell surface antigen CD2 | | Authors: | Evans, E.J, Ikemizu, S, Davis, S.J. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Binding Properties of the CD2 and CD244 (2B4)-binding Protein, CD48
J.Biol.Chem., 281, 2006
|
|
2BBF
 
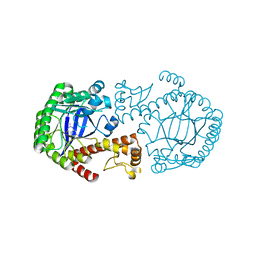 | | Crystal structure of tRNA-guanine transglycosylase (TGT) from Zymomonas mobilis in complex with 6-amino-3,7-dihydro-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-3,7-DIHYDRO-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2005-10-17 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding.
J.Mol.Biol., 370, 2007
|
|
3UL4
 
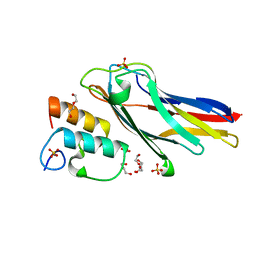 | | Crystal structure of Coh-OlpA(Cthe_3080)-Doc918(Cthe_0918) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome enzyme, dockerin type I, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
2B1A
 
 | | Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with UG1033 peptide | | Descriptor: | Fab 2219, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
J.Virol., 80, 2006
|
|
3T27
 
 | | Orthorhombic trypsin (bovine) in the presence of betaine | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Venkat, M, Saxby, C, Cahn, J, Juers, D. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The use of trimethylamine N-oxide as a primary precipitating agent and related methylamine osmolytes as cryoprotective agents for macromolecular crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2R3Y
 
 | |
2GNC
 
 | | Crystal structure of srGAP1 SH3 domain in the slit-robo signaling pathway | | Descriptor: | SLIT-ROBO Rho GTPase-activating protein 1 | | Authors: | Li, X, Liu, Y, Gao, F, Bartlam, M, Wu, J.Y, Rao, Z. | | Deposit date: | 2006-04-10 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Robo Proline-rich Motif Recognition by the srGAP1 Src Homology 3 Domain in the Slit-Robo Signaling Pathway
J.Biol.Chem., 281, 2006
|
|
2B8R
 
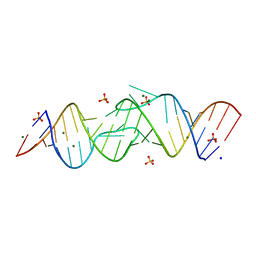 | | Structure oF HIV-1(LAI) genomic RNA DIS | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site
Nat.Struct.Biol., 8, 2001
|
|
1JFW
 
 | |
1JBT
 
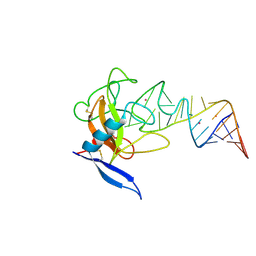 | | CRYSTAL STRUCTURE OF RIBOTOXIN RESTRICTOCIN COMPLEXED WITH A 29-MER SARCIN/RICIN DOMAIN RNA ANALOG | | Descriptor: | 29-MER SARCIN/RICIN DOMAIN RNA ANALOG, POTASSIUM ION, RESTRICTOCIN | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
3T1N
 
 | |
1S17
 
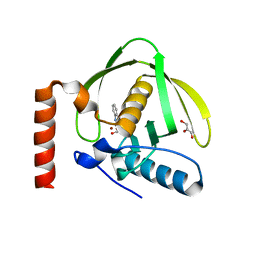 | | Identification of Novel Potent Bicyclic Peptide Deformylase Inhibitors | | Descriptor: | 2-(3,4-DIHYDRO-3-OXO-2H-BENZO[B][1,4]THIAZIN-2-YL)-N-HYDROXYACETAMIDE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Molteni, V, He, X, Nabakka, J, Yang, K, Kreusch, A, Gordon, P, Bursulaya, B, Ryder, N.S, Goldberg, R, He, Y. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of novel potent bicyclic peptide deformylase inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2AS4
 
 | | cytochrome c peroxidase in complex with 3-fluorocatechol | | Descriptor: | 3-FLUOROBENZENE-1,2-DIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Brenk, R, Vetter, S.W, Boyce, S.E, Goodin, D.B, Shoichet, B.K. | | Deposit date: | 2005-08-22 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing molecular docking in a charged model binding site.
J.Mol.Biol., 357, 2006
|
|
2R3U
 
 | | Crystal structure of the PDZ deletion mutant of DegS | | Descriptor: | Protease degS | | Authors: | Clausen, T, Kurzbauer, R. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress.
Genes Dev., 21, 2007
|
|
2GPV
 
 | | Estrogen Related Receptor-gamma ligand binding domain complexed with 4-hydroxy-tamoxifen and a SMRT peptide | | Descriptor: | 4-HYDROXYTAMOXIFEN, Estrogen-related receptor gamma, Nuclear receptor corepressor 2 | | Authors: | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | Deposit date: | 2006-04-18 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
3T26
 
 | | Orthorhombic trypsin (bovine) in the presence of sarcosine | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Cahn, J, Venkat, M, Marshall, H, Juers, D. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The use of trimethylamine N-oxide as a primary precipitating agent and related methylamine osmolytes as cryoprotective agents for macromolecular crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2AZJ
 
 | | Crystal structure for the mutant D81C of Sulfolobus solfataricus hexaprenyl pyrophosphate synthase | | Descriptor: | Geranylgeranyl pyrophosphate synthetase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H.J. | | Deposit date: | 2005-09-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
2B0T
 
 | | Structure of Monomeric NADP Isocitrate dehydrogenase | | Descriptor: | MAGNESIUM ION, NADP Isocitrate dehydrogenase | | Authors: | Imabayashi, F, Aich, S, Prasad, L, Delbaere, L.T. | | Deposit date: | 2005-09-14 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-free structure of a monomeric NADP isocitrate dehydrogenase: An open conformation phylogenetic relationship of isocitrate dehydrogenase.
Proteins, 63, 2006
|
|
