3G5Q
 
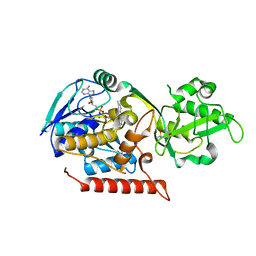 | | Crystal structure of Thermus thermophilus TrmFO | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate--tRNA-(uracil-5-)-methyltransferase trmFO | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5R
 
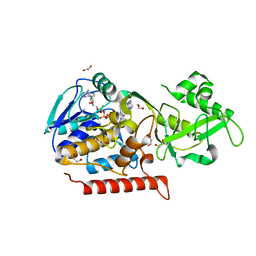 | | Crystal structure of Thermus thermophilus TrmFO in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3OF4
 
 | |
3IWA
 
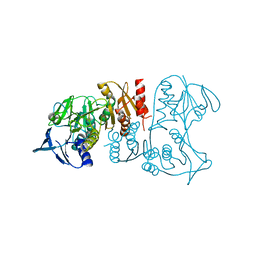 | | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris | | Descriptor: | CALCIUM ION, FAD-dependent pyridine nucleotide-disulphide oxidoreductase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris
To be Published
|
|
1Y56
 
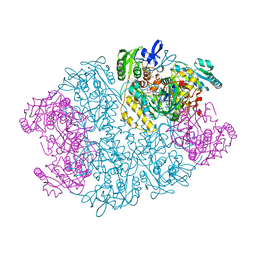 | | Crystal structure of L-proline dehydrogenase from P.horikoshii | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tsuge, H, Kawakami, R, Sakuraba, H, Ago, H, Miyano, M, Aki, K, Katunuma, N, Ohshima, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure of a novel FAD-, FMN-, and ATP-containing L-proline dehydrogenase complex from Pyrococcus horikoshii
J.Biol.Chem., 280, 2005
|
|
3U0I
 
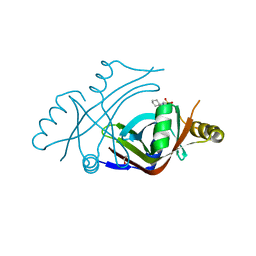 | |
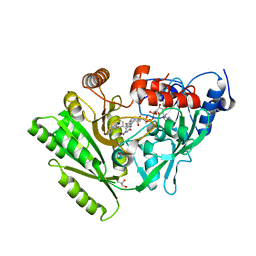
3BW7
 
 | |
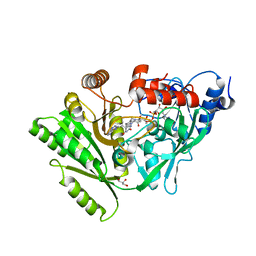
3C0P
 
 | |
8IJY
 
 | | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, Deoxyribodipyrimidine photolyase-related protein, ... | | Authors: | Liu, Y, Xu, L, Zhang, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor
To Be Published
|
|
4XDT
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, a bifunctional FMN transferase/FAD pyrophosphatase, N55Y mutant, FAD bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2014-12-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Evidence for Posttranslational Protein Flavinylation in the Syphilis Spirochete Treponema pallidum: Structural and Biochemical Insights from the Catalytic Core of a Periplasmic Flavin-Trafficking Protein.
Mbio, 6, 2015
|
|
2Z5U
 
 | |
3GDP
 
 | | Hydroxynitrile lyase from almond, monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dreveny, I, Gruber, K, Kratky, C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
8AJK
 
 | | Crystal structure of a C43S variant from the disulfide reductase MerA from Staphylococcus aureus | | Descriptor: | FAD-containing oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Weiland, P, Altegoer, F, Bange, G. | | Deposit date: | 2022-07-28 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MerA functions as a hypothiocyanous acid reductase and defense mechanism in Staphylococcus aureus.
Mol.Microbiol., 119, 2023
|
|
8JZ6
 
 | |
7EIH
 
 | | Ancestral L-Lys oxidase (ligand free form) | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Sugiura, S, Nakano, S, Niwa, M, Hasebe, F, Ito, S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of ancestral L-lysine oxidase assigned by sequence data mining.
J.Biol.Chem., 297, 2021
|
|
7EII
 
 | | Ancestral L-Lys oxidase K387A variant (L-Lys binding form) | | Descriptor: | FAD dependent L-Lys oxidase, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE | | Authors: | Sugiura, S, Nakano, S, Niwa, M, Hasebe, F, Ito, S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic mechanism of ancestral L-lysine oxidase assigned by sequence data mining.
J.Biol.Chem., 297, 2021
|
|
3GDN
 
 | | Almond hydroxynitrile lyase in complex with benzaldehyde | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dreveny, I, Gruber, K, Kratky, C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
4IG1
 
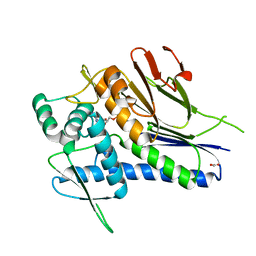 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, Mg(II)-AMP product bound form | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4318 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
5EZ3
 
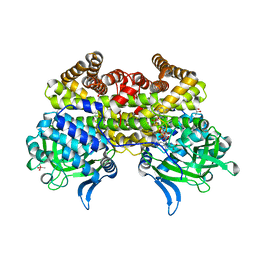 | |
5IDU
 
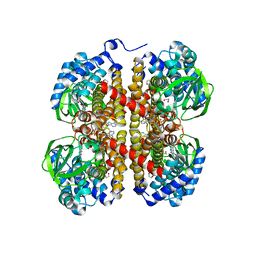 | |
5UFA
 
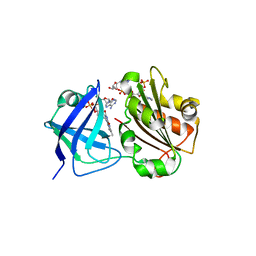 | |
6AWA
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
5U25
 
 | |
5TR9
 
 | |
6AON
 
 | | 1.72 Angstrom Resolution Crystal Structure of 2-Oxoglutarate Dehydrogenase Complex Subunit Dihydrolipoamide Dehydrogenase from Bordetella pertussis in Complex with FAD | | Descriptor: | CALCIUM ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 1.72 Angstrom Resolution Crystal Structure of 2-Oxoglutarate Dehydrogenase Complex Subunit Dihydrolipoamide Dehydrogenase from Bordetella pertussis in Complex with FAD
To Be Published
|
|
