2WZI
 
 | | BtGH84 D243N in complex with 5F-oxazoline | | Descriptor: | (3AS,5S,6S,7R,7AR)-5-FLUORO-5-(HYDROXYMETHYL)-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]OXAZOLE-6,7-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualizing the Reaction Coordinate of an O-Glcnac Hydrolase
J.Am.Chem.Soc., 132, 2010
|
|
6ALO
 
 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and a peroxysuccinate intermediate | | Descriptor: | 4-peroxy-4-oxobutanoic acid, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
2WZP
 
 | | Structures of Lactococcal Phage p2 Baseplate Shed Light on a Novel Mechanism of Host Attachment and Activation in Siphoviridae | | Descriptor: | CAMELID VHH5, LACTOCOCCAL PHAGE P2 ORF15, LACTOCOCCAL PHAGE P2 ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2009-12-01 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6WX6
 
 | |
2WP9
 
 | | Crystal structure of the E. coli succinate:quinone oxidoreductase (SQR) SdhB His207Thr mutant | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Perturbation of the quinone-binding site of complex II alters the electronic properties of the proximal [3Fe-4S] iron-sulfur cluster.
J. Biol. Chem., 286, 2011
|
|
5BZ4
 
 | | Crystal structure of a T1-like thiolase (CoA-complex) from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase, COENZYME A | | Authors: | Janardan, N, Harijan, R.K, Kiema, T.R, Wierenga, R.K, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2EE4
 
 | | Solution structure of the RhoGAP domain from human Rho GTPase activating protein 5 variant | | Descriptor: | Rho GTPase activating protein 5 variant | | Authors: | Tomizawa, T, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RhoGAP domain from human Rho GTPase activating protein 5 variant
To be Published
|
|
2WS0
 
 | | Semi-synthetic analogue of human insulin NMeAlaB26-insulin at pH 7.5 | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6WYU
 
 | | Crystallographic trimer of metal-free TriCyt2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, HEME C, SULFATE ION, ... | | Authors: | Tezcan, F.A, Kakkis, A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4XT9
 
 | | RORgamma (263-509) complexed with GSK2435341A and SRC2 | | Descriptor: | LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, N-[4-(2,5-dichlorophenyl)-5-phenyl-1,3-thiazol-2-yl]-2-[4-(ethylsulfonyl)phenyl]acetamide, Nuclear receptor ROR-gamma, ... | | Authors: | Wang, Y, Ma, Y. | | Deposit date: | 2015-01-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of N-(4-aryl-5-aryloxy-thiazol-2-yl)-amides as potent ROR gamma t inverse agonists
Bioorg.Med.Chem., 23, 2015
|
|
6X0I
 
 | |
5C3G
 
 | | Crystal structure of Bcl-xl bound to BIM-MM | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Bcl-2-like protein 11 | | Authors: | Miles, J.A, Yeo, D.J, Rowell, P, Rodriguez-Marin, S, Pask, C.M, Warriner, S.L, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Chem Sci, 7, 2016
|
|
2EM9
 
 | | Solution structure of the C2H2 type zinc finger (region 367-399) of human Zinc finger protein 224 | | Descriptor: | ZINC ION, Zinc finger protein 224 | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 367-399) of human Zinc finger protein 224
To be Published
|
|
5C1A
 
 | | p97-N750D/R753D/M757D/Q760D in complex with ATP-gamma-S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
6X0J
 
 | |
2EMZ
 
 | | Solution structure of the C2H2 type zinc finger (region 628-660) of human Zinc finger protein 95 homolog | | Descriptor: | ZINC ION, Zinc finger protein 95 homolog | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 628-660) of human Zinc finger protein 95 homolog
To be Published
|
|
2X4S
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a peptide representing the epitope of the H5N1 (Avian Flu) Nucleoprotein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals
J.Am.Chem.Soc., 131, 2009
|
|
2WZJ
 
 | | Catalytic and UBA domain of kinase MARK2/(Par-1) K82R, T208E double mutant | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE MARK2 | | Authors: | Panneerselvam, S, Marx, A, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | Structure and Function of Polarity-Inducing Kinase Family Mark/Par-1 within the Branch of Ampk/Snf1-Related Kinases.
Faseb J., 24, 2010
|
|
6WP2
 
 | |
3FI2
 
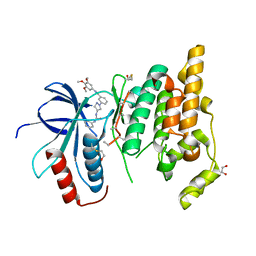 | | Crystal structure of JNK3 with amino-pyrazole inhibitor, SR-3451 | | Descriptor: | 1,2-ETHANEDIOL, 3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}-N-(3,4,5-trimethoxyphenyl)benzamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-activity relationships and X-ray structures describing the selectivity of aminopyrazole inhibitors for c-Jun N-terminal kinase 3 (JNK3) over p38.
J.Biol.Chem., 284, 2009
|
|
1LE9
 
 | | Crystal structure of a NF-kB heterodimer bound to the Ig/HIV-kB siti | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*AP*AP*GP*TP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*T)-3', NUCLEAR FACTOR NF-KAPPA-B P50 SUBUNIT, ... | | Authors: | Benjamin, B, Huang, D.B, Chen-Park, F.E, Sigler, P.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The x-ray crystal structure of the NF-kappa B p50.p65 heterodimer bound to the interferon beta -kappa B site.
J.Biol.Chem., 277, 2002
|
|
6WP9
 
 | | AvaR1 bound to Avenolide | | Descriptor: | (5S)-5-[(6R)-6-hydroxy-6-methyl-5-oxooctyl]furan-2(5H)-one, AvaR1 | | Authors: | Kapoor, I, Olivares, P.J, Nair, S.K. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical basis for the regulation of biosynthesis of antiparasitics by bacterial hormones.
Elife, 9, 2020
|
|
5C8E
 
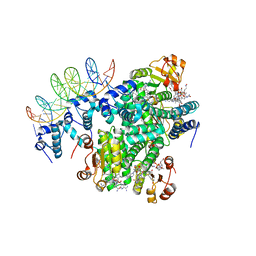 | |
4XXJ
 
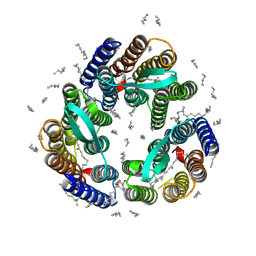 | | Crystal Structure of Escherichia coli-Expressed Halobacterium salinarum Bacteriorhodopsin in the Trimeric Form | | Descriptor: | Bacteriorhodopsin, EICOSANE, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Bratanov, D, Balandin, T, Round, E, Gushchin, I, Gordeliy, V. | | Deposit date: | 2015-01-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Approach to Heterologous Expression of Membrane Proteins. The Case of Bacteriorhodopsin.
Plos One, 10, 2015
|
|
5C69
 
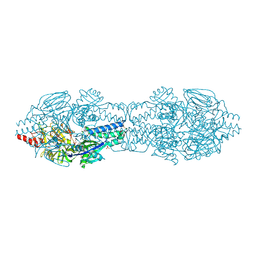 | |
