5B5R
 
 | | Crystal structure of GSDMA3 | | Descriptor: | Gasdermin-A3 | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-15 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Pore-forming activity and structural autoinhibition of the gasdermin family.
Nature, 535, 2016
|
|
5B5Y
 
 | | Crystal structure of PtLCIB4, a homolog of the limiting CO2-inducible protein LCIB | | Descriptor: | ACETATE ION, PtLCIB4, ZINC ION | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Cajar, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
2VFO
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V125L | | Descriptor: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
4Y2U
 
 | | Structure of soluble epoxide hydrolase in complex with tert-butyl 1,2,3,4-tetrahydroquinolin-3-ylcarbamate | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, tert-butyl (3R)-1,2,3,4-tetrahydroquinolin-3-ylcarbamate | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of N-ethylmethylamine as a novel scaffold for inhibitors of soluble epoxide hydrolase by crystallographic fragment screening
Bioorg.Med.Chem., 23, 2015
|
|
1KV7
 
 | | Crystal Structure of CueO, a multi-copper oxidase from E. coli involved in copper homeostasis | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, PROBABLE BLUE-COPPER PROTEIN YACK | | Authors: | Roberts, S.A, Weichsel, A, Grass, G, Thakali, K, Hazzard, J.T, Tollin, G, Rensing, C, Montfort, W.R. | | Deposit date: | 2002-01-25 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3FGO
 
 | | Crystal Structure of the E2 magnesium fluoride complex of the (SR) Ca2+-ATPase with bound CPA and AMPPCP | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Laursen, M, Bublitz, M, Moncoq, K, Olesen, C, Moller, J.V, Young, H.S, Nissen, P, Morth, J.P. | | Deposit date: | 2008-12-08 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cyclopiazonic acid is complexed to a divalent metal ion when bound to the sarcoplasmic reticulum Ca2+-ATPase.
J.Biol.Chem., 2009
|
|
2VLJ
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, Van Der Merwe, A, Bell, J, Mcmichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
4HTH
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+VIAGLA at cryogenic temperature | | Descriptor: | THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Wheeler, E.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B, Robinson, A.C. | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+VIAGLA at cryogenic temperature
To be Published
|
|
5B5P
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with 4-oxo-N-(3-(2-(1H-1,2,4-triazol-3-ylsulfanyl)ethoxy)benzyl)-3,4-dihydroquinazoline-2-carboxamide | | Descriptor: | 4-oxo-N-{3-[2-(1H-1,2,4-triazol-3-ylsulfanyl)ethoxy]benzyl}-3,4-dihydroquinazoline-2-carboxamide, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Novel, Highly Potent, and Selective Matrix Metalloproteinase (MMP)-13 Inhibitors with a 1,2,4-Triazol-3-yl Moiety as a Zinc Binding Group Using a Structure-Based Design Approach
J. Med. Chem., 60, 2017
|
|
1KV2
 
 | | Human p38 MAP Kinase in Complex with BIRB 796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
5B73
 
 | | Crystal structure of human ZMYND8 PHD-Bromo-PWWP domain | | Descriptor: | Protein kinase C-binding protein 1, ZINC ION | | Authors: | Li, H, Li, Y, Zheng, X. | | Deposit date: | 2016-06-03 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
Mol.Cell, 63, 2016
|
|
4Y3D
 
 | |
5B7A
 
 | |
3FI6
 
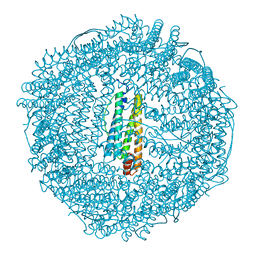 | | apo-H49AFr with high content of Pd ions | | Descriptor: | CADMIUM ION, Ferritin light chain, PALLADIUM ION, ... | | Authors: | Abe, M, Ueno, T, Hirata, K, Suzuki, M, Abe, S, Shimizu, N, Yamaoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2008-12-11 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Process of Accumulation of Metal Ions on the Interior Surface of apo-Ferritin: Crystal Structures of a Series of apo-Ferritins Containing Variable Quantities of Pd(II) Ions
J.Am.Chem.Soc., 131, 2009
|
|
5B6W
 
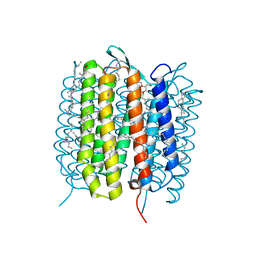 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 16 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
3F8S
 
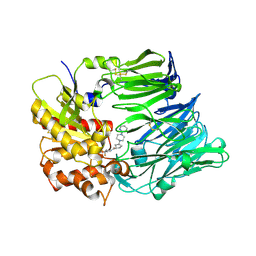 | | Crystal structure of dipeptidyl peptidase IV in complex with inhibitor | | Descriptor: | 2-(4-{(3S,5S)-5-[(3,3-difluoropyrrolidin-1-yl)carbonyl]pyrrolidin-3-yl}piperazin-1-yl)pyrimidine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ammirati, M.J, Liu, S, Piotrowski, D.W. | | Deposit date: | 2008-11-13 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | (3,3-Difluoro-pyrrolidin-1-yl)-[(2S,4S)-(4-(4-pyrimidin-2-yl-piperazin-1-yl)-pyrrolidin-2-yl]-methanone: a potent, selective, orally active dipeptidyl peptidase IV inhibitor.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5B6Z
 
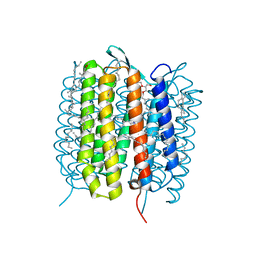 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 1.725 ms us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
2DGN
 
 | | Mouse Muscle Adenylosuccinate Synthetase partially ligated complex with GTP, 2'-deoxy-IMP | | Descriptor: | 9-(2-DEOXY-5-O-PHOSPHONO-BETA-D-ERYTHRO-PENTOFURANOSYL)-6-(PHOSPHONOOXY)-9H-PURINE, Adenylosuccinate synthetase isozyme 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Zhou, Y, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cavitation as a mechanism of substrate discrimination by adenylosuccinate synthetases.
Biochemistry, 45, 2006
|
|
1KTD
 
 | | CRYSTAL STRUCTURE OF CLASS II MHC MOLECULE IEK BOUND TO PIGEON CYTOCHROME C PEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion protein consisting of cytochrome C peptide, glycine rich linker, ... | | Authors: | Fremont, D.H, Dai, S, Chiang, H, Crawford, F, Marrack, P, Kappler, J. | | Deposit date: | 2002-01-15 | | Release date: | 2002-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of cytochrome c presentation by IE(k).
J.Exp.Med., 195, 2002
|
|
3FIA
 
 | | Crystal structure of the EH 1 domain from human intersectin-1 protein. Northeast Structural Genomics Consortium target HR3646e. | | Descriptor: | CALCIUM ION, Intersectin-1, SULFATE ION | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Devices, C, Zhao, L, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-11 | | Release date: | 2008-12-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the EH 1 domain from human intersectin-1 protein.
To be Published
|
|
4XOA
 
 | | Crystal structure of a FimH*DsG complex from E.coli K12 in space group P1 | | Descriptor: | FimG, Protein FimH | | Authors: | Jakob, R.P, Eras, J, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
3FIL
 
 | |
4HWK
 
 | | Crystal structure of human sepiapterin reductase in complex with sulfapyridine | | Descriptor: | 4-amino-N-(pyridin-2-yl)benzenesulfonamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Groenlund Pedersen, M, Pojer, F, Johnsson, K. | | Deposit date: | 2012-11-08 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tetrahydrobiopterin biosynthesis as an off-target of sulfa drugs.
Science, 340, 2013
|
|
6A7X
 
 | | Rat Xanthine oxidoreductase, D428A variant, NAD bound form | | Descriptor: | BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
3FJ7
 
 | | Crystal structure of L-phospholactate Bound PEB3 | | Descriptor: | L-PHOSPHOLACTATE, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
