5TXW
 
 | |
8R3B
 
 | |
6H5D
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 2 | | Descriptor: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
7RCQ
 
 | |
7M5I
 
 | | Endolysin from Escherichia coli O157:H7 phage FAHEc1 | | Descriptor: | Endolysin, PHOSPHATE ION | | Authors: | Love, M.J, Coombes, D, Billington, C, Dobson, R.C.J. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Molecular Basis for Escherichia coli O157:H7 Phage FAHEc1 Endolysin Function and Protein Engineering to Increase Thermal Stability.
Viruses, 13, 2021
|
|
7RH0
 
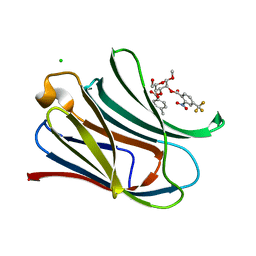 | |
5TY2
 
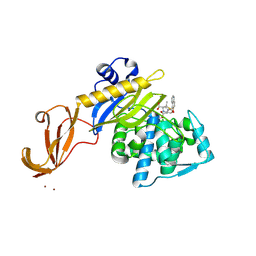 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-18 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
6DD8
 
 | | Structure of mouse SYCP3, P21 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
5HY2
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
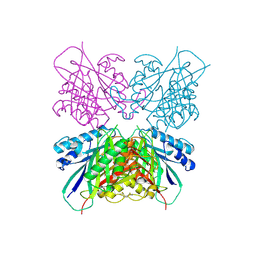
7MDS
 
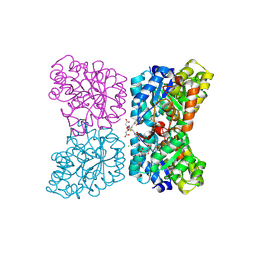 | | Crystal structure of AtDHDPS1 in complex with MBDTA-2 | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase 1, chloroplastic, CHLORIDE ION, ... | | Authors: | Hall, C.J, Soares da Costa, T.P, Panjikar, S. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Towards novel herbicide modes of action by inhibiting lysine biosynthesis in plants.
Elife, 10, 2021
|
|
5TYI
 
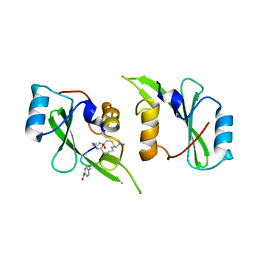 | | Grb7 SH2 with bicyclic peptide containing pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, Peptide inhibitor | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
8PM8
 
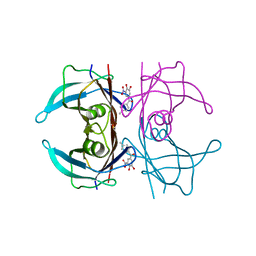 | | V30M Transthyretin structure in complex with Tolcalpone | | Descriptor: | Tolcapone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
8PO6
 
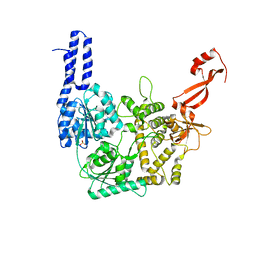 | | Structure of Escherichia coli HrpA apo form | | Descriptor: | ATP-dependent RNA helicase HrpA, PHOSPHATE ION | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
6IGP
 
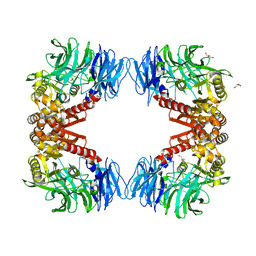 | | Crystal structure of S9 peptidase (inactive state)from Deinococcus radiodurans R1 in P212121 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
7R1G
 
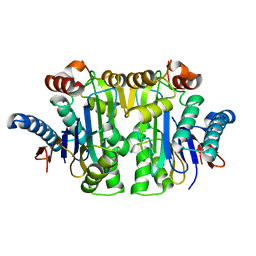 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-38 (R207C, D210S, S211V) | | Descriptor: | Beta-aspartyl-peptidase, Isoaspartyl peptidase, SODIUM ION | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8PM9
 
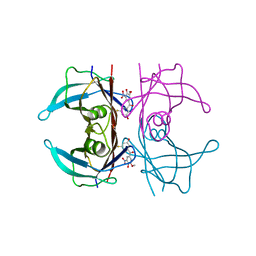 | | Crystal structure of human wild type transthyretin in complex with PITB (Pharmacokinetically Improved TTR Binder) | | Descriptor: | (3-fluoranyl-5-oxidanyl-phenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
6IJE
 
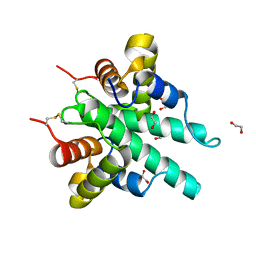 | | Crystal structure of the type VI amidase immunity (Tai4) from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, Tai4 | | Authors: | Fukuhara, S, Nakane, T, Yamashita, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2018-10-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the Agrobacterium tumefaciens type VI effector-immunity complex.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6GJD
 
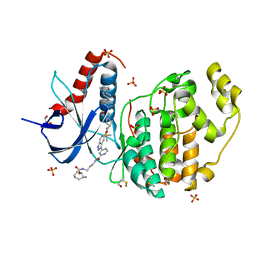 | | Erk2 signalling protein | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | O'Reilly, M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Quantitation of ERK1/2 inhibitor cellular target occupancies with a reversible slow off-rate probe.
Chem Sci, 9, 2018
|
|
8PZ8
 
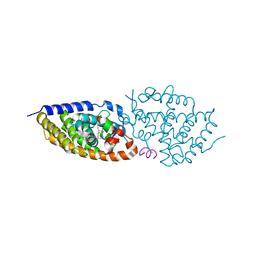 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 54 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
5TZJ
 
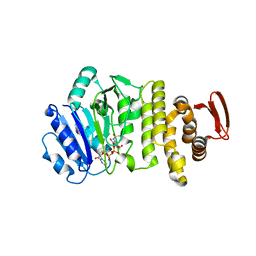 | | Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
6GJN
 
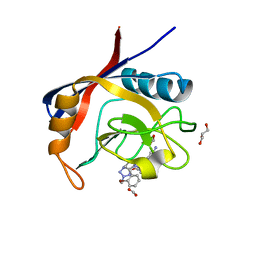 | | Cyclophilin A complexed with tri-vector ligand 15. | | Descriptor: | 1-[(4-aminophenyl)methyl]-3-[2-[(2~{R})-2-(2-bromophenyl)pyrrolidin-1-yl]-2-oxidanylidene-ethyl]-1-[(2-methyl-1,2,3,4-tetrazol-5-yl)methyl]urea, FORMIC ACID, GLYCEROL, ... | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6DOR
 
 | |
8PO7
 
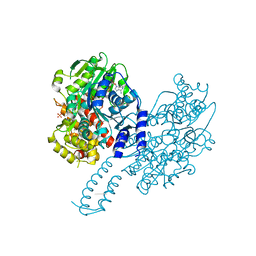 | | Structure of Escherichia coli HrpA in complex with ADP and dinucleotide dCdC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
6DF2
 
 | |
7LVW
 
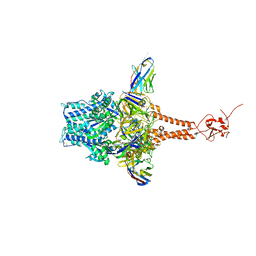 | | Structure of RSV F in Complex with VHH Cl184 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, F-VHH-Cl184, ... | | Authors: | Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2021-02-26 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A vulnerable, membrane-proximal site in human respiratory syncytial virus F revealed by a prefusion-specific single-domain antibody.
J.Virol., 95, 2021
|
|
