1BVV
 
 | |
3WUM
 
 | |
3WUX
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, Unsaturated chondroitin disaccharide hydrolase | | Authors: | Nakamichi, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae
to be published
|
|
1C3V
 
 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
3O69
 
 | | Structure of the E100A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GDP-mannose pyrophosphatase nudK, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
1L7X
 
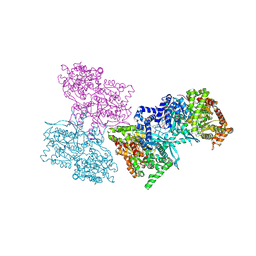 | | Human liver glycogen phosphorylase b complexed with caffeine, N-acetyl-beta-D-glucopyranosylamine, and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CAFFEINE, Glycogen phosphorylase, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-18 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
3WPK
 
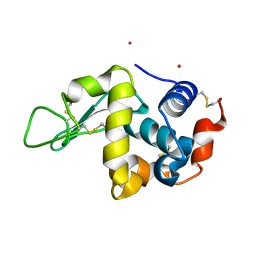 | |
3A22
 
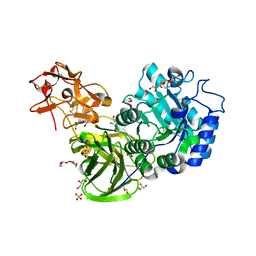 | | Crystal Structure of beta-L-Arabinopyranosidase complexed with L-arabinose | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2009-04-27 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.
J.Biol.Chem., 284, 2009
|
|
1BWI
 
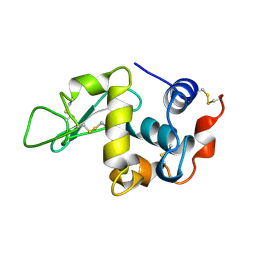 | | THE 1.8 A STRUCTURE OF MICROBATCH OIL DROP GROWN TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME) | | Authors: | Dong, J, Boggon, T.J, Chayen, N.E, Raftery, J, Bi, R.C. | | Deposit date: | 1998-09-24 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bound-solvent structures for microgravity-, ground control-, gel- and microbatch-grown hen egg-white lysozyme crystals at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6ADL
 
 | |
3A67
 
 | | Crystal Structure of HyHEL-10 Fv mutant LN31D complexed with hen egg white lysozyme | | Descriptor: | IG VH, anti-lysozyme, Lysozyme C, ... | | Authors: | Yokota, A, Tsumoto, K, Shiroishi, M, Nakanishi, T, Kondo, H, Kumagai, I. | | Deposit date: | 2009-08-24 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of asparagine residues to the stabilization of a proteinaceous antigen-antibody complex, HyHEL-10-hen egg white lysozyme
J.Biol.Chem., 285, 2010
|
|
3WWN
 
 | | Crystal structure of LysZ from Thermus thermophilus complex with LysW | | Descriptor: | OrfF, Putative acetylglutamate kinase-like protein, SULFATE ION, ... | | Authors: | Yoshida, A, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2014-06-21 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into amino group-carrier protein-mediated lysine biosynthesis: crystal structure of the LysZ·LysW complex from Thermus thermophilus.
J.Biol.Chem., 290, 2015
|
|
7XKY
 
 | |
1FHT
 
 | | RNA-BINDING DOMAIN OF THE U1A SPLICEOSOMAL PROTEIN U1A117, NMR, 43 STRUCTURES | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1996-02-21 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding.
J.Mol.Biol., 257, 1996
|
|
3A46
 
 | | Crystal structure of MvNei1/THF complex | | Descriptor: | DNA, Formamidopyrimidine-DNA glycosylase, GLYCEROL | | Authors: | Imamura, K, Wallace, S, Doublie, S. | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of a Viral NEIL1 Ortholog Unliganded and Bound to Abasic Site-containing DNA
J.Biol.Chem., 284, 2009
|
|
3WRM
 
 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of P450cam intermedite
To be published
|
|
3A6J
 
 | | E122Q mutant creatininase complexed with creatine | | Descriptor: | Creatinine amidohydrolase, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
4HTN
 
 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 1.32 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WX0
 
 | |
3OCD
 
 | | Diheme SoxAX - C236M mutant | | Descriptor: | HEME C, SoxA, SoxX | | Authors: | Maher, M.J. | | Deposit date: | 2010-08-09 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diheme SoxAX proteins - insights into structure and function of the active site
To be Published
|
|
1BYA
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
7XBR
 
 | | Crystal structure of phosphorylated AtMKK5 | | Descriptor: | Mitogen-activated protein kinase kinase 5 | | Authors: | Pei, C.J, Luo, Z.P, Wu, J.W, Wang, Z.X. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the phosphorylated Arabidopsis MKK5 reveals activation mechanism of MAPK kinases.
Acta Biochim.Biophys.Sin., 54, 2022
|
|
3W7P
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with TT2-4-031 | | Descriptor: | 5-[2-(4-carboxyphenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with TT2-4-031
To be Published
|
|
3WSP
 
 | | Crystal Structure of P450BM3 with N-perfluorononanoyl-L-tryptophan | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, DIMETHYL SULFOXIDE, N-(2,2,3,3,4,4,5,5,6,6,7,7,8,8,9,9,9-heptadecafluorononanoyl)-L-tryptophan, ... | | Authors: | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2014-03-20 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of Wild-type Cytochrome P450BM3 by the Next Generation of Decoy Molecules: Enhanced Hydroxylation of Gaseous Alkanes and Crystallographic Evidence.
ACS CATALYSIS, 5, 2015
|
|
3W88
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-200 | | Descriptor: | 1,2-ETHANEDIOL, 5-[4-(6-carboxynaphthalen-2-yl)butyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, CACODYLATE ION, ... | | Authors: | Inaoka, D.K, Hashimoto, S, Rocha, J.R, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Kuranaga, T, Shiba, T, Balogun, E.O, Sakamoto, K, Suzuki, S, Montanari, C.A, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-200
To be Published
|
|
