4HHS
 
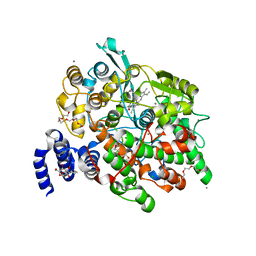 | | Crystal Structure of fatty acid alpha-dioxygenase (Arabidopsis thaliana) | | Descriptor: | Alpha-dioxygenase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Goulah, C.C, Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2012-10-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of alpha-Dioxygenase provides insight into diversity in the cyclooxygenase-peroxidase superfamily.
Biochemistry, 52, 2013
|
|
1TSN
 
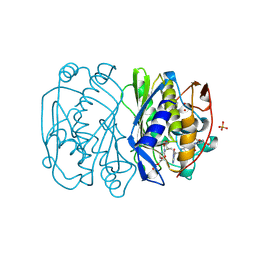 | | THYMIDYLATE SYNTHASE TERNARY COMPLEX WITH FDUMP AND METHYLENETETRAHYDROFOLATE | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, PHOSPHATE ION, ... | | Authors: | Hyatt, D.C, Maley, F, Montfort, W.R. | | Deposit date: | 1996-12-02 | | Release date: | 1997-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Use of strain in a stereospecific catalytic mechanism: crystal structures of Escherichia coli thymidylate synthase bound to FdUMP and methylenetetrahydrofolate.
Biochemistry, 36, 1997
|
|
1U5B
 
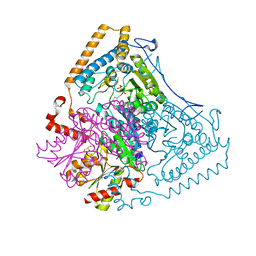 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, GLYCEROL, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-07-27 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
4I4B
 
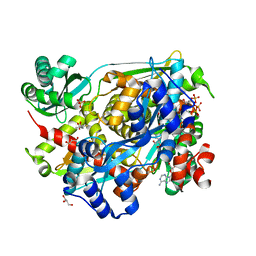 | | HMG-CoA Reductase from Pseudomonas mevalonii complexed with NAD and intermediate hemiacetal form of HMG-CoA | | Descriptor: | (3R,5R,9R,19R,21S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,21-tetrahydroxy-8,8,21-trimethyl-10,14-dioxo-19-sulfanyl-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide, (3S)-3-hydroxy-3-methyl-5-sulfanylpentanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, ... | | Authors: | Stauffacher, C.V, Steussy, C.N, Critchelow, C.J, Schmidt, T, Min, J, Wrensford, L.V, Burgner, J.W, Rodwell, V.W. | | Deposit date: | 2012-11-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
2V5P
 
 | | COMPLEX STRUCTURE OF HUMAN IGF2R DOMAINS 11-13 BOUND TO IGF-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, INSULIN-LIKE GROWTH FACTOR II, ... | | Authors: | Brown, J, Delaine, C, Zaccheo, O.J, Siebold, C, Gilbert, R.J, van Boxel, G, Denley, A, Wallace, J.C, Hassan, A.B, Forbes, B.E, Jones, E.Y. | | Deposit date: | 2007-07-06 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Embo J., 27, 2008
|
|
4I56
 
 | | HMG-CoA reductase from pseudomonas mevalonii complexed with dithio-HMG-coa | | Descriptor: | (3S,5S,9R,21S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,21-tetrahydroxy-8,8,21-trimethyl-10,14-dioxo-19-thioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, ... | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Crichelow, C.J, Rodwell, V.W, Wrensford, L.V, Min, J, Burgner II, J.W. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
3W1Q
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with NL-2 | | Descriptor: | 5-(3,3-dimethylbutyl)-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-20 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with NL-2
To be Published
|
|
4HK9
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2Z27
 
 | | Thr109Ser dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
3O2N
 
 | |
1F6W
 
 | |
1U8U
 
 | | E. coli Thioesterase I/Protease I/Lysophospholiase L1 in complexed with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, GLYCEROL, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Lo, Y.-C, Lin, S.-C, Liaw, Y.-C. | | Deposit date: | 2004-08-07 | | Release date: | 2005-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Substrate specificities of Escherichia coli thioesterase I/protease I/lysophospholipase L1 are governed by its switch loop movement
Biochemistry, 44, 2005
|
|
1FGM
 
 | |
8A9Q
 
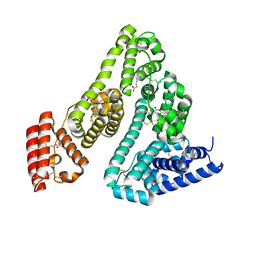 | | Computational design of stable mammalian serum albumins for bacterial expression | | Descriptor: | Albumin, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Khersonsky, O, Dym, O, Fleishman, J.S. | | Deposit date: | 2022-06-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stable Mammalian Serum Albumins Designed for Bacterial Expression.
J.Mol.Biol., 435, 2023
|
|
2ZWB
 
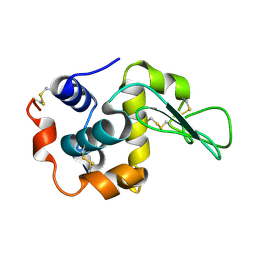 | | Neutron crystal structure of wild type human lysozyme in D2O | | Descriptor: | Lysozyme C | | Authors: | Chiba-Kamoshida, K, Matsui, T, Chatake, T, Ohhara, T, Ostermann, A, Tanaka, I, Yutani, K, Niimura, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Site-specific softening of peptide bonds by localized deuterium observed by neutron crystallography of human lysozyme
To be Published
|
|
3W2K
 
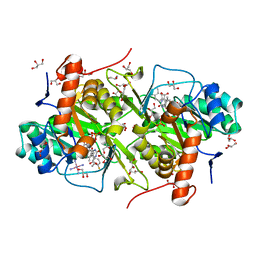 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-165 | | Descriptor: | 5-[2-(4-fluoro-3-methylphenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-29 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-165
To be Published
|
|
1FHL
 
 | |
3W2N
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-185 | | Descriptor: | 2,6-dioxo-5-{2-[2-(trifluoromethyl)phenyl]ethyl}-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-185
To be Published
|
|
1BDM
 
 | |
5ZRT
 
 | | Crystal structure of human C1ORF123 protein | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rahaman, S.N.A, Yusop, J.M, Mohamed-Hussein, Z.A, Wan Mohd, A, Ho, K.L, Teh, A.H, Waterman, J, Ng, C.L. | | Deposit date: | 2018-04-25 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of human C1ORF123.
Peerj, 6, 2018
|
|
1KQN
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
4I7R
 
 | |
3OEA
 
 | | Crystal structure of the Q121E mutants of C.polysaccharolyticus CBM16-1 bound to cellopentaose | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mutational insights into the roles of amino acid residues in ligand binding for two closely related family 16 carbohydrate binding modules.
J.Biol.Chem., 285, 2010
|
|
3W2E
 
 | | Crystal structure of oxidation intermediate (20 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
2V6X
 
 | | Stractural insight into the interaction between ESCRT-III and Vps4 | | Descriptor: | DOA4-INDEPENDENT DEGRADATION PROTEIN 4, SULFATE ION, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 4 | | Authors: | Obita, T, Perisic, O, Ghazi-Tabatabai, S, Saksena, S, Emr, S.D, Williams, R.L. | | Deposit date: | 2007-07-23 | | Release date: | 2007-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis for Selective Recognition of Escrt-III by the Aaa ATPase Vps4
Nature, 449, 2007
|
|
