4OAQ
 
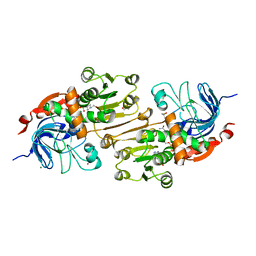 | | Crystal structure of the R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Aggrawal, N, Mandal, P.K, Gautham, N, Chadha, A. | | Deposit date: | 2014-01-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Insights into the stereoselectivity of R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330: Biochemical Characterization and Crystal structure studies
To be Published
|
|
4K1P
 
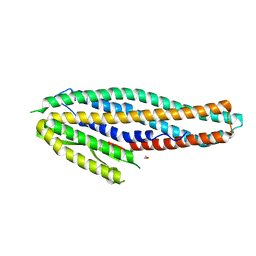 | | Structure of the NheA component of the Nhe toxin from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, NheA, SULFATE ION | | Authors: | Ganash, M, Phung, D, Artymiuk, P.J. | | Deposit date: | 2013-04-05 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the NheA Component of the Nhe Toxin from Bacillus cereus: Implications for Function.
Plos One, 8, 2013
|
|
1OQJ
 
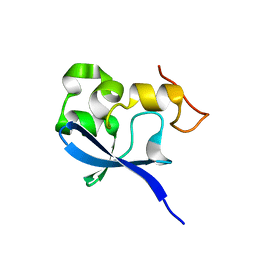 | | Crystal structure of the SAND domain from glucocorticoid modulatory element binding protein-1 (GMEB1) | | Descriptor: | Glucocorticoid Modulatory Element Binding protein-1, ZINC ION | | Authors: | Surdo, P.L, Bottomley, M.J, Sattler, M, Scheffzek, K. | | Deposit date: | 2003-03-10 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucocorticoid modulatory element binding protein-1 reveals deoxyribonucleic acid and zinc binding regions
MOL.ENDOCRINOL., 17, 2003
|
|
1OVN
 
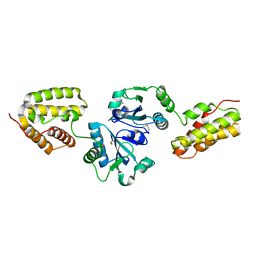 | | Crystal Structure and Functional Analysis of Drosophila Wind-- a PDI-Related Protein | | Descriptor: | CESIUM ION, Windbeutel | | Authors: | Ma, Q, Guo, C, Barnewitz, K, Sheldrick, G.M, Soling, H.D, Uson, I, Ferrari, D.M. | | Deposit date: | 2003-03-27 | | Release date: | 2004-02-24 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of Drosophila Wind, a protein-disulfide isomerase-related protein.
J.Biol.Chem., 278, 2003
|
|
4L04
 
 | |
4F8P
 
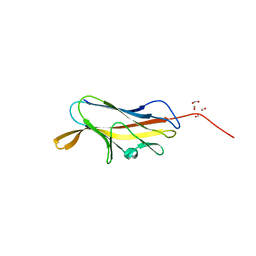 | | X-ray structure of PsaA from Yersinia pestis, in complex with galactose | | Descriptor: | ACETATE ION, TERT-BUTYL FORMATE, beta-D-galactopyranose, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L1Z
 
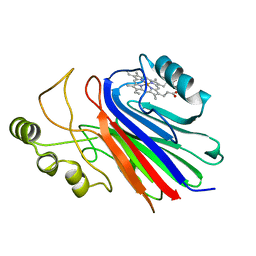 | |
1TCJ
 
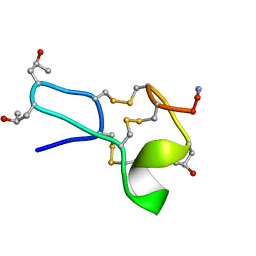 | |
1THG
 
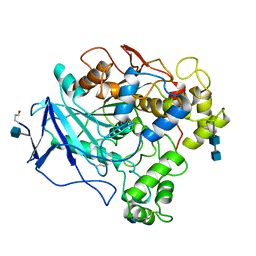 | | 1.8 ANGSTROMS REFINED STRUCTURE OF THE LIPASE FROM GEOTRICHUM CANDIDUM | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schrag, J.D, Cygler, M. | | Deposit date: | 1992-07-28 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A refined structure of the lipase from Geotrichum candidum.
J.Mol.Biol., 230, 1993
|
|
4IGG
 
 | |
4KZO
 
 | |
3O37
 
 | |
1MST
 
 | |
1ITU
 
 | | HUMAN RENAL DIPEPTIDASE COMPLEXED WITH CILASTATIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CILASTATIN, RENAL DIPEPTIDASE, ... | | Authors: | Nitanai, Y, Satow, Y, Adachi, H, Tsujimoto, M. | | Deposit date: | 2002-02-03 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Renal Dipeptidase Involved in beta-Lactam Hydrolysis
J.Mol.Biol., 321, 2002
|
|
1SY9
 
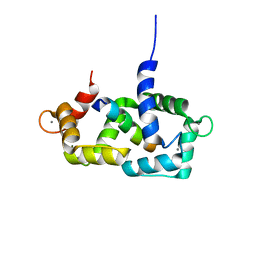 | | Structure of calmodulin complexed with a fragment of the olfactory CNG channel | | Descriptor: | CALCIUM ION, CALMODULIN, Cyclic-nucleotide-gated olfactory channel | | Authors: | Contessa, G.M, Orsale, M, Melino, S, Torre, V, Paci, M, Desideri, A, Cicero, D.O. | | Deposit date: | 2004-04-01 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of calmodulin complexed with an olfactory CNG channel fragment and role of the central linker: residual dipolar couplings to evaluate calmodulin binding modes outside the kinase family.
J.Biomol.Nmr, 31, 2005
|
|
4L06
 
 | |
1TF6
 
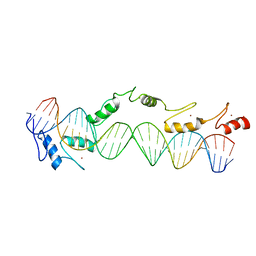 | | CO-CRYSTAL STRUCTURE OF XENOPUS TFIIIA ZINC FINGER DOMAIN BOUND TO THE 5S RIBOSOMAL RNA GENE INTERNAL CONTROL REGION | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*GP*CP*CP*TP*GP*GP*TP*TP*AP*GP*TP*AP*C P*CP*TP*GP*GP*AP* TP*GP*GP*GP*AP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*TP*CP*CP*CP*AP*TP*CP*CP*AP*GP*GP*T P*AP*CP*TP*AP*AP* CP*CP*AP*GP*GP*CP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR IIIA), ... | | Authors: | Nolte, R.T, Conlin, R.M, Harrison, S.C, Brown, R.S. | | Deposit date: | 1998-03-02 | | Release date: | 1998-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1DCI
 
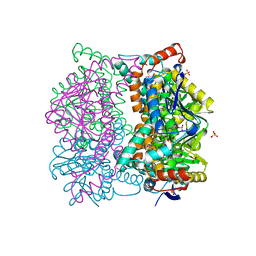 | | DIENOYL-COA ISOMERASE | | Descriptor: | 1,2-ETHANEDIOL, DIENOYL-COA ISOMERASE, MAGNESIUM ION, ... | | Authors: | Modis, Y, Filppula, S.A, Novikov, D, Norledge, B, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 1998-02-13 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of dienoyl-CoA isomerase at 1.5 A resolution reveals the importance of aspartate and glutamate sidechains for catalysis.
Structure, 6, 1998
|
|
1OZV
 
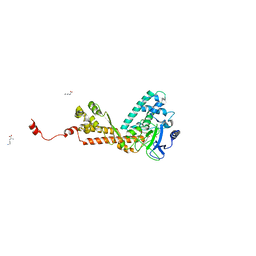 | | Crystal structure of the SET domain of LSMT bound to Lysine and AdoHcy | | Descriptor: | LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast, ... | | Authors: | Trievel, R.C, Flynn, E.M, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2003-04-09 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT
Nat.Struct.Biol., 10, 2003
|
|
1IDS
 
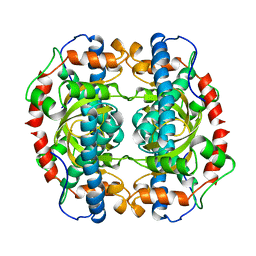 | | X-RAY STRUCTURE ANALYSIS OF THE IRON-DEPENDENT SUPEROXIDE DISMUTASE FROM MYCOBACTERIUM TUBERCULOSIS AT 2.0 ANGSTROMS RESOLUTIONS REVEALS NOVEL DIMER-DIMER INTERACTIONS | | Descriptor: | FE (III) ION, IRON SUPEROXIDE DISMUTASE | | Authors: | Cooper, J.B, Mcintyre, K, Wood, S.P, Zhang, Y, Young, D. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of the iron-dependent superoxide dismutase from Mycobacterium tuberculosis at 2.0 Angstroms resolution reveals novel dimer-dimer interactions.
J.Mol.Biol., 246, 1995
|
|
4CMW
 
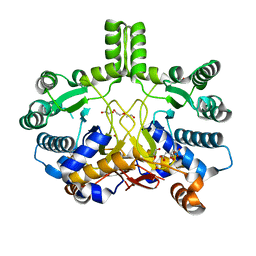 | | Crystal structure of Rv3378c | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Layre, E, Lee, H.J, Young, D.C, Martinot, A.J, Buter, J, Minnaard, A.J, Annand, J.W, Fortune, S.M, Snider, B.B, Matsunaga, I, Rubin, E.J, Alber, T, Moody, D.B. | | Deposit date: | 2014-01-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Molecular Profiling of Mycobacterium Tuberculosis Identifies Tuberculosinyl Nucleoside Products of the Virulence-Associated Enzyme Rv3378C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZHG
 
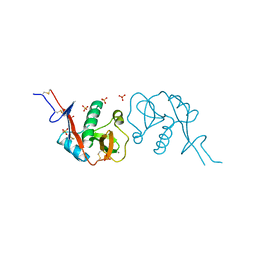 | |
2ADZ
 
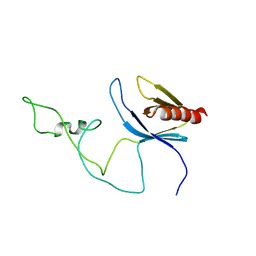 | | solution structure of the joined PH domain of alpha1-syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Wen, W, Xu, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1P8H
 
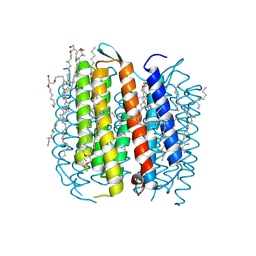 | | BACTERIORHODOPSIN M1 INTERMEDIATE PRODUCED AT ROOM TEMPERATURE | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, Bacteriorhodopsin, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic Structures of the M and N Intermediates of Bacteriorhodopsin: Assembly of a Hydrogen-Bonded Chain of Water Molecules between Asp-96 and the Retinal Schiff Base
J.Mol.Biol., 330, 2003
|
|
1DAW
 
 | | CRYSTAL STRUCTURE OF A BINARY COMPLEX OF PROTEIN KINASE CK2 (ALPHA-SUBUNIT) AND MG-AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN KINASE CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.G, Schomburg, D. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GTP plus water mimic ATP in the active site of protein kinase CK2.
Nat.Struct.Biol., 6, 1999
|
|
