1HY3
 
 | | CRYSTAL STRUCTURE OF HUMAN ESTROGEN SULFOTRANSFERASE V269E MUTANT IN THE PRESENCE OF PAPS | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, ESTROGEN SULFOTRANSFERASE | | Authors: | Pedersen, L.C, Petrochenko, E.V, Shevtsov, S, Negishi, M. | | Deposit date: | 2001-01-17 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction.
J.Biol.Chem., 277, 2002
|
|
1Q67
 
 | | Crystal structure of Dcp1p | | Descriptor: | Decapping protein involved in mRNA degradation-Dcp1p | | Authors: | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | Deposit date: | 2003-08-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PRB
 
 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN PAB | | Authors: | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
1PFM
 
 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 1-15 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
1PFN
 
 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 16-27 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
3ZHZ
 
 | | Structure of Mycobacterium tuberculosis DXR in complex with a fosmidomycin analogue | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, [(1S)-1-(3,4-dichlorophenyl)-3-[oxidanyl-[2-[[3-(trifluoromethyl)phenyl]amino]phenyl]carbonyl-amino]propyl]phosphonic acid | | Authors: | Bjorkelid, C, Jansson, A.M, Bergfors, T, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2012-12-30 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dxr Inhibition by Potent Mono- and Disubstituted Fosmidomycin Analogues.
J.Med.Chem., 56, 2013
|
|
4N64
 
 | |
4D4G
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D57
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, APNA A1, ARGININE, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4CMX
 
 | | Crystal structure of Rv3378c | | Descriptor: | 1,2-ETHANEDIOL, BENZENE HEXACARBOXYLIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Layre, E, Lee, H.J, Young, D.C, Martinot, A.J, Buter, J, Minnaard, A.J, Annand, J.W, Fortune, S.M, Snider, B.B, Matsunaga, I, Rubin, E.J, Alber, T, Moody, D.B. | | Deposit date: | 2014-01-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular Profiling of Mycobacterium Tuberculosis Identifies Tuberculosinyl Nucleoside Products of the Virulence-Associated Enzyme Rv3378C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OG5
 
 | | Human menin with bound inhibitor MIV-5 | | Descriptor: | 4-(3-{4-[(S)-cyclopentyl(hydroxy)pyridin-2-ylmethyl]piperidin-1-yl}propoxy)benzonitrile, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | He, S, Senter, T.J, Pollock, J.W, Han, C, Upadhyay, S.K, Purohit, T, Gogliotti, R.D, Lindsley, C.W, Cierpicki, T, Stauffer, S.R, Grembecka, J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | High-Affinity Small-Molecule Inhibitors of the Menin-Mixed Lineage Leukemia (MLL) Interaction Closely Mimic a Natural Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
4D4H
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, GLYCEROL | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1RF5
 
 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in Unliganded State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
1RI4
 
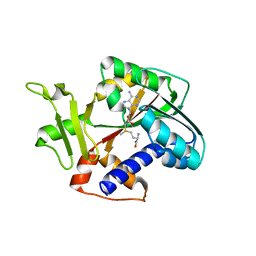 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RIW
 
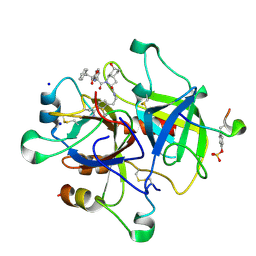 | | Thrombin in complex with natural product inhibitor Oscillarin | | Descriptor: | (2R,3AS,6R,7AS)-N-(2-{1-[AMINO(IMINO)METHYL]-2,5-DIHYDRO-1H-PYRROL-3-YL}ETHYL)-6-HYDROXY-1-{N-[(2S)-2-HYDROXY-3-PHENYLPROPANOYL]PHENYLALANYL}OCTAHYDRO-1H-INDOLE-2-CARBOXAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin IIB, ... | | Authors: | Hanessian, S, Tremblay, M, Petersen, J.F.W. | | Deposit date: | 2003-11-18 | | Release date: | 2004-11-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The N-acyloxyiminium ion aza-Prins route to octahydroindoles: total synthesis and structural confirmation of the antithrombotic marine natural product oscillarin
J.Am.Chem.Soc., 126, 2004
|
|
1RI2
 
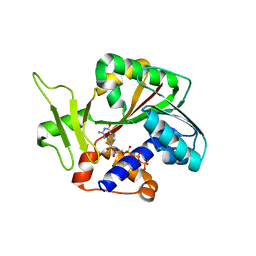 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
4OG4
 
 | | Human menin with bound inhibitor MIV-3S | | Descriptor: | 4-(3-{4-[(S)-cyclopentyl(hydroxy)phenylmethyl]piperidin-1-yl}propoxy)benzonitrile, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | He, S, Senter, T.J, Pollock, J.W, Han, C, Upadhyay, S.K, Purohit, T, Gogliotti, R.D, Lindsley, C.W, Cierpicki, T, Stauffer, S.R, Grembecka, J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Affinity Small-Molecule Inhibitors of the Menin-Mixed Lineage Leukemia (MLL) Interaction Closely Mimic a Natural Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
1JLC
 
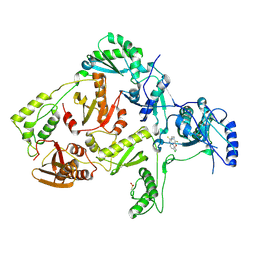 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | Descriptor: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1MAE
 
 | |
1RRU
 
 | | The influence of a chiral amino acid on the helical handedness of PNA in solution and in crystals | | Descriptor: | Peptide Nucleic Acid, (H-P(*CPN*GPN*TPN*APN*CPN*GPN)-LYS-NH2) | | Authors: | Rasmussen, H, Liljefors, T, Petersson, B, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Influence of a Chiral Amino Acid on the Helical Handedness of PNA in Solution and in Crystals
J.Biomol.Struct.Dyn., 21, 2004
|
|
3O2M
 
 | |
3O33
 
 | |
1MSY
 
 | |
3O4K
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) and lipoteichoic acid at 2.1 A resolution | | Descriptor: | (2S)-1-({3-O-[2-(acetylamino)-4-amino-2,4,6-trideoxy-beta-D-galactopyranosyl]-alpha-D-glucopyranosyl}oxy)-3-(heptanoyloxy)propan-2-yl (7Z)-pentadec-7-enoate, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of recognition of pathogen-associated molecular patterns and inhibition of proinflammatory cytokines by camel peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3X1L
 
 | |
