1ACX
 
 | |
196L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
198L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
197L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
2VU8
 
 | |
1CTN
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE A | | Authors: | Perrakis, A, Tews, I, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial chitinase at 2.3 A resolution.
Structure, 2, 1994
|
|
199L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
2K96
 
 | | Solution structure of the RDC-refined P2B-P3 pseudoknot from human telomerase RNA (delta U177) | | Descriptor: | TELOMERASE RNA P2B-P3 PSEUDOKNOT | | Authors: | Kim, N.-K, Zhang, Q, Zhou, J, Theimer, C.A, Peterson, R.D, Feigon, J. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Wild-type Pseudoknot of Human Telomerase RNA.
J.Mol.Biol., 384, 2008
|
|
2LPR
 
 | |
2JTC
 
 | | 3D structure and backbone dynamics of SPE B | | Descriptor: | Streptopain | | Authors: | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | Deposit date: | 2007-07-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
1SBT
 
 | | ATOMIC COORDINATES FOR SUBTILISIN BPN (OR NOVO) | | Descriptor: | SUBTILISIN BPN' | | Authors: | Alden, R.A, Birktoft, J.J, Kraut, J, Robertus, J.D, Wright, C.S. | | Deposit date: | 1972-08-11 | | Release date: | 1977-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic coordinates for subtilisin BPN' (or Novo).
Biochem.Biophys.Res.Commun., 45, 1971
|
|
1FI8
 
 | | RAT GRANZYME B [N66Q] COMPLEXED TO ECOTIN [81-84 IEPD] | | Descriptor: | ECOTIN, NATURAL KILLER CELL PROTEASE 1 | | Authors: | Waugh, S.M, Harris, J.L, Fletterick, R.J, Craik, C.S. | | Deposit date: | 2000-08-03 | | Release date: | 2000-09-13 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the pro-apoptotic protease granzyme B reveals the molecular determinants of its specificity
Nat.Struct.Biol., 7, 2000
|
|
3EST
 
 | | STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, PORCINE PANCREATIC ELASTASE, SULFATE ION | | Authors: | Meyer, E.F, Cole, G, Radhakrishnan, R, Epp, O. | | Deposit date: | 1987-09-17 | | Release date: | 1988-01-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of native porcine pancreatic elastase at 1.65 A resolutions.
Acta Crystallogr.,Sect.B, 44, 1988
|
|
1UMA
 
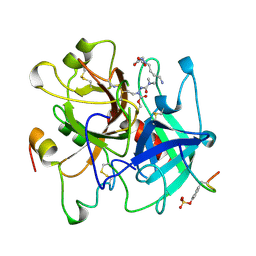 | | ALPHA-THROMBIN (HIRUGEN) COMPLEXED WITH NA-(N,N-DIMETHYLCARBAMOYL)-ALPHA-AZALYSINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUDIN I, ... | | Authors: | Nardini, M, Pesce, A, Rizzi, M, Casale, E, Ferraccioli, R, Balliano, G, Milla, P, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1996-03-26 | | Release date: | 1996-11-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the active site titrant N alpha-(N,N-dimethylcarbamoyl)-alpha-azalysine p-nitrophenyl ester: a comparative kinetic and X-ray crystallographic study.
J.Mol.Biol., 258, 1996
|
|
1WIP
 
 | |
3HTC
 
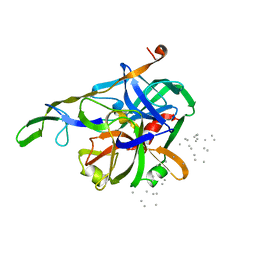 | | THE STRUCTURE OF A COMPLEX OF RECOMBINANT HIRUDIN AND HUMAN ALPHA-THROMBIN | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUDIN VARIANT 2 | | Authors: | Tulinsky, A, Rydel, T.J, Ravichandran, K.G, Huber, R, Bode, W. | | Deposit date: | 1993-06-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a complex of recombinant hirudin and human alpha-thrombin.
Science, 249, 1990
|
|
1EYG
 
 | |
1AD3
 
 | |
2V77
 
 | | Crystal Structure of Human Carboxypeptidase A1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARBOXYPEPTIDASE A1, OCTANE-1,3,5,7-TETRACARBOXYLIC ACID, ... | | Authors: | Pallares, I, Fernandez, D, Comellas-Bigler, M, Fernandez-Recio, J, Ventura, S, Aviles, F.X, Vendrell, J. | | Deposit date: | 2007-07-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Direct interaction between a human digestive protease and the mucoadhesive poly(acrylic acid).
Acta Crystallogr. D Biol. Crystallogr., D64, 2008
|
|
1GQF
 
 | |
1HT0
 
 | | HUMAN GAMMA-2 ALCOHOL DEHYDROGENSE | | Descriptor: | CLASS I ALCOHOL DEHYDROGENASE 3, GAMMA SUBUNIT, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Niederhut, M.S, Gibbons, B.J, Perez-Miller, S, Hurley, T.D. | | Deposit date: | 2000-12-27 | | Release date: | 2001-01-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of the three human class I alcohol dehydrogenases.
Protein Sci., 10, 2001
|
|
3I2A
 
 | |
3LUM
 
 | | Structure of ulilysin mutant M290L | | Descriptor: | ARGININE, CALCIUM ION, GLYCEROL, ... | | Authors: | Tallant, C, Garcia-Castellanos, R, Baumann, U, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | On the relevance of the Met-turn methionine in metzincins.
J.Biol.Chem., 285, 2010
|
|
3HGP
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by high resolution crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
3HGN
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by neutron crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (1.65 Å), X-RAY DIFFRACTION | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
