4WQ8
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - tetrathionate soak | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.3989 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
1Y4J
 
 | | Crystal structure of the paralogue of the human formylglycine generating enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dickmanns, A, Rudolph, M.G, Ficner, R. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Crystal Structure of Human pFGE, the Paralog of the C{alpha}-formylglycine-generating Enzyme
J.Biol.Chem., 280, 2005
|
|
1L3Q
 
 | | H. rufescens abalone shell Lustrin A consensus repeat, FPGKNVNCTSGE, pH 7.4, 1-H NMR structure | | Descriptor: | Lustrin A | | Authors: | Evans, J.S, Wustman, B.A, Zhang, B, Morse, D.E. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Model peptide studies of sequence regions in the elastomeric biomineralization protein, Lustrin A. I. The C-domain consensus-PG-, -NVNCT-motif
Biopolymers, 63, 2002
|
|
2XME
 
 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE, GLYCEROL | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Archaeoglobus Fulgidus Ctp:Inositol-1-Phosphate Cytidylyltransferase, a Key Enzyme for Di-Myo-Inositol-Phosphate Synthesis in (Hyper)Thermophiles.
J.Bacteriol., 193, 2011
|
|
5UYW
 
 | | YfeA ancillary sites that co-load with site 2 | | Descriptor: | Periplasmic chelated iron-binding protein YfeA, ZINC ION | | Authors: | Radka, C.D, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of Yersinia pestis virulence factor YfeA reveals two polyspecific metal-binding sites.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4X60
 
 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and sinefungin | | Descriptor: | GLYCEROL, Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
4WPK
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form I | | Descriptor: | CITRIC ACID, SODIUM ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3GJ7
 
 | |
1D32
 
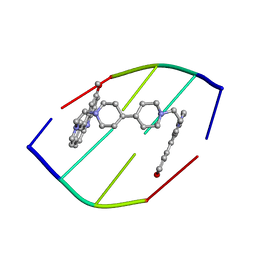 | | DRUG-INDUCED DNA REPAIR: X-RAY STRUCTURE OF A DNA-DITERCALINIUM COMPLEX | | Descriptor: | DITERCALINIUM, DNA (5'-D(*CP*GP*CP*G)-3') | | Authors: | Gao, Q, Williams, L.D, Egli, M, Rabinovich, D, Chen, S.-L, Quigley, G.J, Rich, A. | | Deposit date: | 1991-01-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Drug-induced DNA repair: X-ray structure of a DNA-ditercalinium complex.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
3GJ4
 
 | |
4X61
 
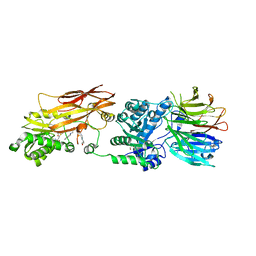 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and SAM | | Descriptor: | GLYCEROL, Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
2BOB
 
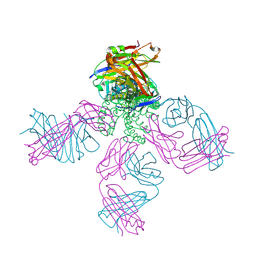 | | Potassium channel KcsA-Fab complex in thallium with tetrabutylammonium (TBA) | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, COBALT (II) ION, ... | | Authors: | Lenaeus, M.J, Vamvouka, M, Focia, P.J, Gross, A. | | Deposit date: | 2005-04-09 | | Release date: | 2005-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Basis of Tea Blockade in a Model Potassium Channel
Nat.Struct.Mol.Biol., 12, 2005
|
|
3KIO
 
 | | mouse RNase H2 complex | | Descriptor: | Ribonuclease H2 subunit A, Ribonuclease H2 subunit B, Ribonuclease H2 subunit C | | Authors: | Shaban, N, Harvey, S, Perrino, F.W, Hollis, T. | | Deposit date: | 2009-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the mammalian RNase H2 complex provides insight into RNA:DNA hybrid processing to prevent immune dysfunction.
J.Biol.Chem., 285, 2010
|
|
4WV3
 
 | | Crystal structure of the anthranilate CoA ligase AuaEII in complex with anthranoyl-AMP | | Descriptor: | 5'-O-[(S)-[(2-aminobenzoyl)oxy](hydroxy)phosphoryl]adenosine, Anthranilate-CoA ligase | | Authors: | Tsai, S.C, Muller, R, Jackson, D.R, Tu, S.S, Nguyen, M, Barajas, J.F, Pistorious, D, Krug, D. | | Deposit date: | 2014-11-04 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structural Insights into Anthranilate Priming during Type II Polyketide Biosynthesis.
Acs Chem.Biol., 11, 2016
|
|
6IKA
 
 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:entecavir-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6J2J
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with MERS-CoV-derived peptide MERS-CoV-S3 | | Descriptor: | Beta-2-microglobulin, MERS-CoV-S3, Ptal-N*01:01 | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
3G81
 
 | |
5VXG
 
 | | Crystal structure of Xanthomonas campestris OleA E117Q bound with Cerulenin | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | Authors: | Jensen, M.R, Wilmot, C.M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | OleA Glu117 is key to condensation of two fatty-acyl coenzyme A substrates in long-chain olefin biosynthesis.
Biochem. J., 474, 2017
|
|
6RNW
 
 | |
3GJ5
 
 | |
4ZAK
 
 | | Crystal structure of the mCD1d/DB06-1/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Birkholz, A.M. | | Deposit date: | 2015-04-13 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.824 Å) | | Cite: | A Novel Glycolipid Antigen for NKT Cells That Preferentially Induces IFN-gamma Production.
J Immunol., 195, 2015
|
|
6RTM
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 2 hour of soaking | | Descriptor: | 1-methylidenenaphthalen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
3QEX
 
 | |
6BT4
 
 | | Crystal structure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit | | Descriptor: | 2-(acetylamino)-4-O-{2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranose, S-layer protein sap, SULFATE ION | | Authors: | Sychantha, D, Chapman, R.N, Bamford, N.C, Boons, G.J, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-12-05 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Molecular Basis for the Attachment of S-Layer Proteins to the Cell Wall of Bacillus anthracis.
Biochemistry, 57, 2018
|
|
6RZK
 
 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:3(Chlorine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3-chlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.046 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
