1FFT
 
 | | The structure of ubiquinol oxidase from Escherichia coli | | Descriptor: | COPPER (II) ION, HEME O, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abramson, J, Riistama, S, Larsson, G, Jasaitis, A, Svensson-Ek, M, Puustinen, A, Iwata, S, Wikstrom, M. | | Deposit date: | 2000-07-26 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site.
Nat.Struct.Biol., 7, 2000
|
|
7S9A
 
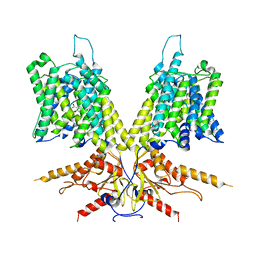 | | Cryo-EM Structure of dolphin Prestin: Inhibited I (Chloride + Salicylate) | | Descriptor: | 2-HYDROXYBENZOIC ACID, Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9B
 
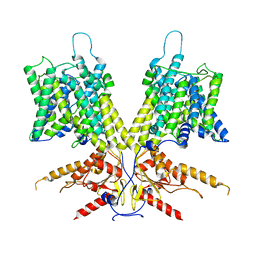 | | Cryo-EM Structure of dolphin Prestin: Sensor Down I (Expanded) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
2C8X
 
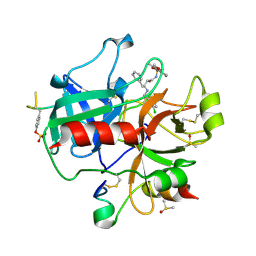 | | thrombin inhibitors | | Descriptor: | DIMETHYL SULFOXIDE, HIRUDIN VARIANT-2, N-{(2R,3S)-3-[(3-CHLOROBENZYL)AMINO]-2-HYDROXY-4-PHENYLBUTYL}-4-METHOXY-2,3,6-TRIMETHYLBENZENESULFONAMIDE, ... | | Authors: | Howard, N, Abell, C, Blakemore, W, Carr, R, Chessari, G, Congreve, M, Howard, S, Jhoti, H, Murray, C.W, Seavers, L.C.A, van Montfort, R.L.M. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Application of Fragment Screening and Fragment Linking to the Discovery of Novel Thrombin Inhibitors
J.Med.Chem., 49, 2006
|
|
1FO6
 
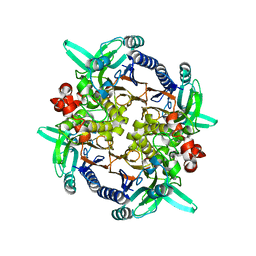 | | CRYSTAL STRUCTURE ANALYSIS OF N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE | | Descriptor: | N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE, XENON | | Authors: | Wang, W.-C, Hsu, W.-H, Chien, F.-T, Chen, C.-Y. | | Deposit date: | 2000-08-25 | | Release date: | 2001-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and site-directed mutagenesis studies of N-carbamoyl-D-amino-acid amidohydrolase from Agrobacterium radiobacter reveals a homotetramer and insight into a catalytic cleft.
J.Mol.Biol., 306, 2001
|
|
1A0S
 
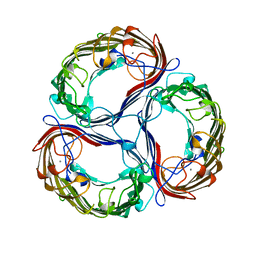 | | SUCROSE-SPECIFIC PORIN | | Descriptor: | CALCIUM ION, SUCROSE-SPECIFIC PORIN | | Authors: | Diederichs, K, Welte, W. | | Deposit date: | 1997-12-07 | | Release date: | 1998-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose.
Nat.Struct.Biol., 5, 1998
|
|
1FOB
 
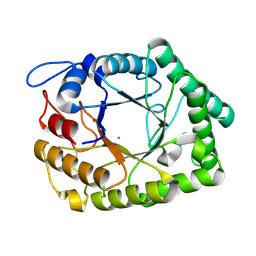 | |
1FQZ
 
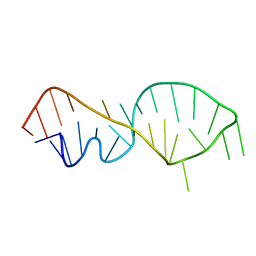 | | NMR VALIDATED MODEL OF DOMAIN IIID OF HEPATITIS C VIRUS INTERNAL RIBOSOME ENTRY SITE | | Descriptor: | HEPATITIS C VIRUS IRES DOMAIN IIID | | Authors: | Klinck, R, Westhof, E, Walker, S, Afshar, M, Collier, A, Aboul-ela, F. | | Deposit date: | 2000-09-07 | | Release date: | 2001-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A potential RNA drug target in the hepatitis C virus internal ribosomal entry site.
RNA, 6, 2000
|
|
1AH6
 
 | |
2WVF
 
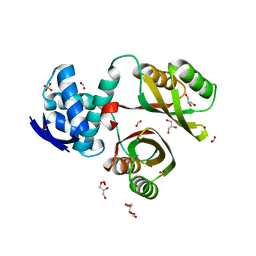 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | FORMIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
1A0T
 
 | | SUCROSE-SPECIFIC PORIN, WITH BOUND SUCROSE MOLECULES | | Descriptor: | CALCIUM ION, SUCROSE-SPECIFIC PORIN, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Diederichs, K, Welte, W. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose.
Nat.Struct.Biol., 5, 1998
|
|
2HBV
 
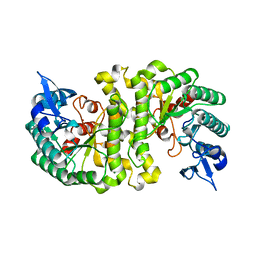 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
1OUB
 
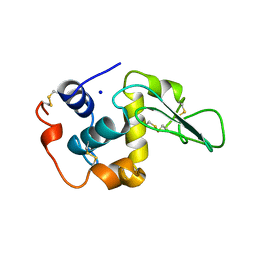 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V100A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1HIG
 
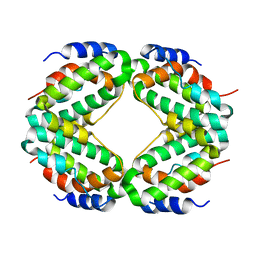 | | THREE-DIMENSIONAL STRUCTURE OF RECOMBINANT HUMAN INTERFERON-GAMMA. | | Descriptor: | INTERFERON-GAMMA | | Authors: | Ealick, S.E, Cook, W.J, Vijay-Kumar, S, Carson, M, Nagabhushan, T.L, Trotta, P.P, Bugg, C.E. | | Deposit date: | 1991-10-03 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Three-dimensional structure of recombinant human interferon-gamma.
Science, 252, 1991
|
|
1A2X
 
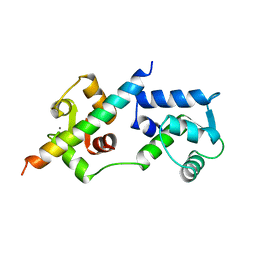 | | COMPLEX OF TROPONIN C WITH A 47 RESIDUE (1-47) FRAGMENT OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, TROPONIN I | | Authors: | Vassylyev, D.G, Takeda, S, Wakatsuki, S, Maeda, K, Maeda, Y. | | Deposit date: | 1998-01-13 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of troponin C in complex with troponin I fragment at 2.3-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8SY8
 
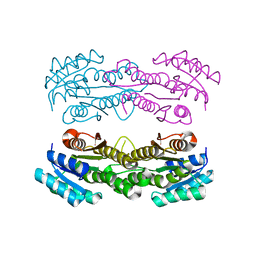 | | Crystal structure of TsaC | | Descriptor: | 4-formylbenzenesulfonate dehydrogenase TsaC | | Authors: | Boggs, D.G, Tian, J, Bridwell-Rabb, J. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The NADH recycling enzymes TsaC and TsaD regenerate reducing equivalents for Rieske oxygenase chemistry.
J.Biol.Chem., 299, 2023
|
|
1AMY
 
 | |
1AN7
 
 | |
1AA0
 
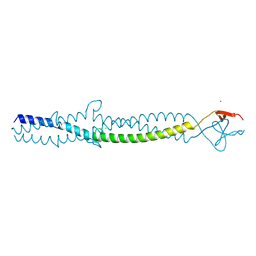 | | FIBRITIN DELETION MUTANT E (BACTERIOPHAGE T4) | | Descriptor: | CHLORIDE ION, FIBRITIN, ZINC ION | | Authors: | Tao, Y, Strelkov, S.V, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 1997-01-18 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain.
Structure, 5, 1997
|
|
8SZO
 
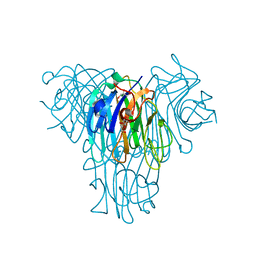 | | Canavalia villosa lectin in complex with alpha-methyl-mannoside | | Descriptor: | CALCIUM ION, Canavalia villosa lectin, GLYCEROL, ... | | Authors: | Cavada, B.S, Lossio, C.F, Pinto-Junior, V.R, Osterne, V.J.S, Oliveira, M.V, Neco, A.H.B, Nascimento, K.S. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lectin from Canavalia villosa seeds: A glucose/mannose-specific protein and a new tool for inflammation studies.
Int J Biol Macromol, 105, 2017
|
|
1AH8
 
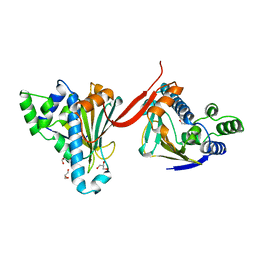 | |
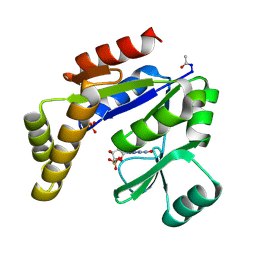
1GKY
 
 | |
1A62
 
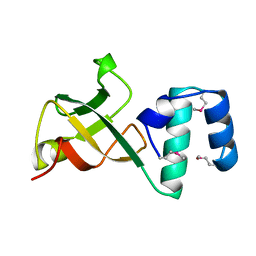 | | CRYSTAL STRUCTURE OF THE RNA-BINDING DOMAIN OF THE TRANSCRIPTIONAL TERMINATOR PROTEIN RHO | | Descriptor: | RHO | | Authors: | Allison, T.J, Wood, T.C, Briercheck, D.M, Rastinejad, F, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-06-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the RNA-binding domain from transcription termination factor rho.
Nat.Struct.Biol., 5, 1998
|
|
1ABA
 
 | | THE STRUCTURE OF OXIDIZED BACTERIOPHAGE T4 GLUTAREDOXIN (THIOREDOXIN). REFINEMENT OF NATIVE AND MUTANT PROTEINS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTAREDOXIN | | Authors: | Eklund, H, Ingelman, M, Soderberg, B.-O, Uhlin, T, Nordlund, P, Nikkola, M, Sonnerstam, U, Joelson, T, Petratos, K. | | Deposit date: | 1992-04-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of oxidized bacteriophage T4 glutaredoxin (thioredoxin). Refinement of native and mutant proteins.
J.Mol.Biol., 228, 1992
|
|
1AAZ
 
 | | THE STRUCTURE OF OXIDIZED BACTERIOPHAGE T4 GLUTAREDOXIN (THIOREDOXIN) | | Descriptor: | CADMIUM ION, GLUTAREDOXIN | | Authors: | Eklund, H, Ingelman, M, Soderberg, B.-O, Uhlin, T, Nordlund, P, Nikkola, M, Sonnerstam, U, Joelson, T, Petratos, K. | | Deposit date: | 1992-04-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of oxidized bacteriophage T4 glutaredoxin (thioredoxin). Refinement of native and mutant proteins.
J.Mol.Biol., 228, 1992
|
|
