4OX0
 
 | |
1NAW
 
 | | ENOLPYRUVYL TRANSFERASE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | Authors: | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | Deposit date: | 1996-07-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
4OXI
 
 | | Crystal structure of Vibrio cholerae adenylation domain AlmE in complex with glycyl-adenosine-5'-phosphate | | Descriptor: | Enterobactin synthetase component F-related protein, GLYCYL-ADENOSINE-5'-PHOSPHATE | | Authors: | Fage, C.D, Henderson, J.C, Keatinge-Clay, A.T, Trent, M.S. | | Deposit date: | 2014-02-05 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Antimicrobial peptide resistance of Vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases.
Acs Chem.Biol., 9, 2014
|
|
1NB9
 
 | | Crystal Structure of Riboflavin Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOFLAVIN, ... | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
4OY7
 
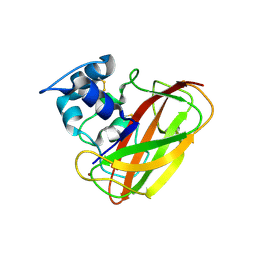 | | Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Putative secreted cellulose binding protein | | Authors: | Forsberg, Z, Mackenzie, A.K, Sorlie, M, Rohr, A.K, Helland, R, Arvai, A.S, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2014-02-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1NBL
 
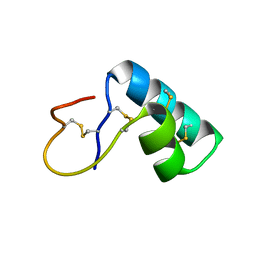 | |
4OZA
 
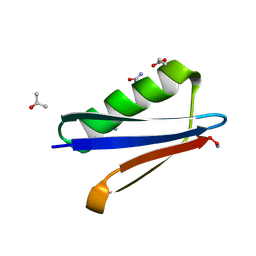 | | Backbone Modifications in the Protein GB1 Helix: beta-3-Ala24, beta-3-Lys28, beta-3-Gln32, beta-3-Asp36 | | Descriptor: | ISOPROPYL ALCOHOL, Streptococcal Protein GB1 Backbone Modified Variant: beta-3-Ala24, beta-3-Lys28, ... | | Authors: | Reinert, Z.E, Horne, W.S. | | Deposit date: | 2014-02-14 | | Release date: | 2014-07-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Folding Thermodynamics of Protein-Like Oligomers with Heterogeneous Backbones.
Chem Sci, 5, 2014
|
|
1NC0
 
 | |
4P00
 
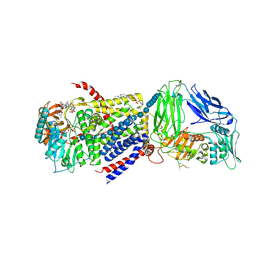 | | Bacterial Cellulose Synthase in complex with cyclic-di-GMP and UDP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Cellulose Synthase A subunit, ... | | Authors: | Morgan, J.L.W, McNamara, J.T, Zimmer, J. | | Deposit date: | 2014-02-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1NBJ
 
 | | High-resolution solution structure of cycloviolacin O1 | | Descriptor: | cycloviolacin O1 | | Authors: | Rosengren, K.J, Daly, N.L, Plan, M.R, Waine, C, Craik, D.J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Twists, Knots, and Rings in Proteins. STRUCTURAL DEFINITION OF THE CYCLOTIDE FRAMEWORK.
J.Biol.Chem., 278, 2003
|
|
4OVO
 
 | |
1NCH
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
4P10
 
 | | pro-carboxypeptidase U In Complex With 5-(3-aminopropyl)-1-propyl-6,7-dihydro-4H-benzimidazole-5-carboxylic acid | | Descriptor: | (5R)-5-(3-aminopropyl)-1-propyl-4,5,6,7-tetrahydro-1H-benzimidazole-5-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase B2, ... | | Authors: | Hallberg, K, Sjogren, T. | | Deposit date: | 2014-02-21 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of conformationally restricted inhibitors of active thrombin activatable fibrinolysis inhibitor (TAFIa).
Bioorg.Med.Chem., 22, 2014
|
|
1NBR
 
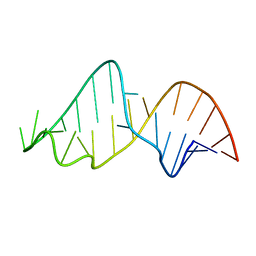 | |
4P1O
 
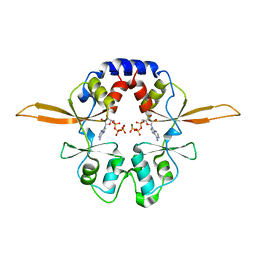 | | Crystal structure of the Bateman domain of murine magnesium transporter CNNM2 bound to ATP-Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Abascal-Palacios, G, Diercks, T, Oyenarte, I, Ereno-Orbea, J, Encinar, J.A, Spiwok, V, Terashima, H, Accardi, A, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To Be Published
|
|
1NCA
 
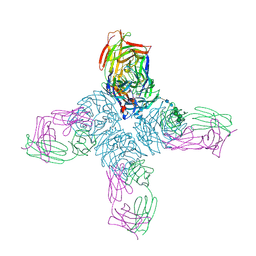 | | REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex.
J.Mol.Biol., 227, 1992
|
|
4P1L
 
 | | Crystal structure of a trap periplasmic solute binding protein from chromohalobacter salexigens dsm 3043 (csal_2479), target EFI-510085, with bound d-glucuronate, spg i213 | | Descriptor: | GLYCEROL, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1NDY
 
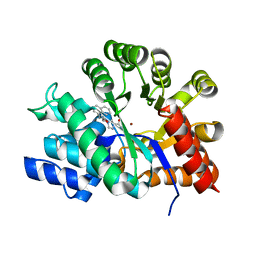 | | Crystal Structure of Adenosine Deaminase Complexed with FR230513 | | Descriptor: | 1-((1R)-1-(HYDROXYMETHYL)-3-(1-NAPHTHYL)PROPYL)-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine Deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2002-12-09 | | Release date: | 2003-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A highly potent non-nucleoside adenosine deaminase inhibitor: efficient drug discovery by intentional lead hybridization
J.Am.Chem.Soc., 126, 2004
|
|
4P1W
 
 | |
1NEC
 
 | | NITROREDUCTASE FROM ENTEROBACTER CLOACAE | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (NITROREDUCTASE) | | Authors: | Hecht, H.J, Bryant, C, Erdmann, H, Pelletier, H, Sawaya, R. | | Deposit date: | 1999-03-30 | | Release date: | 2000-03-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Nitroreductase from Enterobacter Cloacae
To be Published
|
|
4P2I
 
 | | Crystal structure of the mouse SNX19 PX domain | | Descriptor: | MKIAA0254 protein | | Authors: | Collins, B.M. | | Deposit date: | 2014-03-04 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Different Phosphoinositide Specificities of the PX Domains of Sorting Nexins Regulating G-protein Signaling.
J.Biol.Chem., 289, 2014
|
|
1ND0
 
 | |
4P2Q
 
 | | Crystal structure of the 5cc7 TCR in complex with 5c2/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5c2 peptide, 5cc7 T-cell receptor alpha chain, ... | | Authors: | Birnbaum, M.E, Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-03-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Deconstructing the Peptide-MHC Specificity of T Cell Recognition.
Cell, 157, 2014
|
|
1NEV
 
 | | A-tract decamer | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*CP*GP*G)-3' | | Authors: | Barbic, A, Zimmer, D.P, Crothers, D.M. | | Deposit date: | 2002-12-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural origins of adenine-tract bending
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4P4K
 
 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between allergic hypersensitivity and auto immunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM, HLA class II histocompatibility antigen, ... | | Authors: | Clayton, G.M, Crawford, F, Kappler, J.W. | | Deposit date: | 2014-03-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
