1BQ3
 
 | |
3ZXW
 
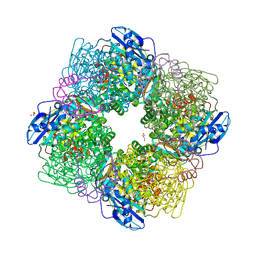 | | STRUCTURE OF ACTIVATED RUBISCO FROM THERMOSYNECHOCOCCUS ELONGATUS COMPLEXED WITH 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Terlecka, B, Wilhelmi, V, Bialek, W, Gubernator, B, Szczepaniak, A, Hofmann, E. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase from Thermosynechococcus Elongatus
To be Published
|
|
5W1K
 
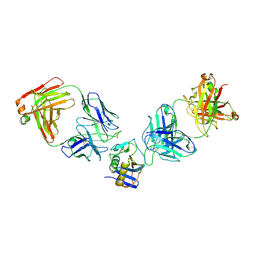 | | JUNV GP1 CR1-10 Fab CR1-28 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR1-10 Fab heavy chain, ... | | Authors: | Raymond, D.D, Clark, L.E, Abraham, J. | | Deposit date: | 2017-06-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses.
Nat Commun, 9, 2018
|
|
5L4I
 
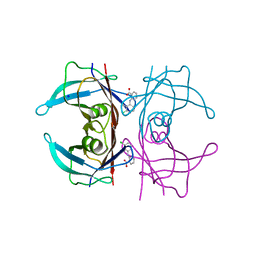 | | Crystal Structure of Human Transthyretin in Complex with Clonixin | | Descriptor: | 2-(3-chloro-2-methylanilino)pyridine-3-carboxylic acid, SODIUM ION, Transthyretin | | Authors: | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
5NTB
 
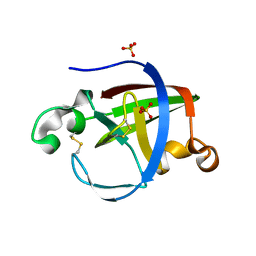 | | Streptomyces papain inhibitor (SPI) | | Descriptor: | Papain inhibitor, SULFATE ION | | Authors: | Schmelz, S, Juettner, N.E, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Illuminating structure and acyl donor sites of a physiological transglutaminase substrate from Streptomyces mobaraensis.
Protein Sci., 27, 2018
|
|
1BQH
 
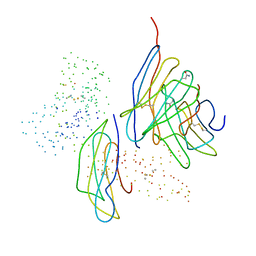 | | MURINE CD8AA ECTODOMAIN FRAGMENT IN COMPLEX WITH H-2KB/VSV8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (BETA-2-MICROGLOBULIN ), PROTEIN (CD8A OR LYT2 OR LYT-2), ... | | Authors: | Wang, J.H, Reinherz, E.L, Kern, P.S, Chang, H.C. | | Deposit date: | 1998-08-16 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of CD8 coreceptor function revealed by crystallographic analysis of a murine CD8alphaalpha ectodomain fragment in complex with H-2Kb.
Immunity, 9, 1998
|
|
7W6E
 
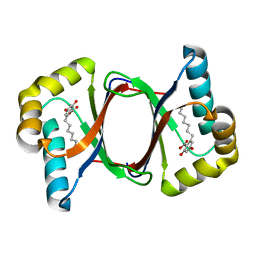 | |
6II0
 
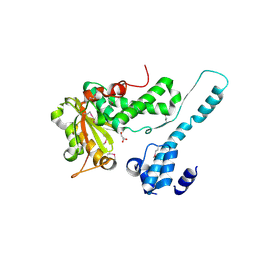 | | Crystal structure of the Makes Caterpillars Floppy (MCF)-Like effector of Vibrio vulnificus MO6-24/O | | Descriptor: | GLYCEROL, Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyun, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2LC9
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
1RTI
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 1-(2-HYDROXYETHYLOXYMETHYL)-6-PHENYL THIOTHYMINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
3ZD4
 
 | |
1JI5
 
 | | Dlp-1 from bacillus anthracis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dlp-1, FE (III) ION | | Authors: | Papinutto, E, Dundon, W.G, Pitulis, N, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of two iron-binding proteins from Bacillus anthracis.
J.Biol.Chem., 277, 2002
|
|
8W2G
 
 | | Human liver phosphofructokinase-1 in the R-state conformation | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B.A. | | Deposit date: | 2024-02-20 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for allosteric regulation of human phosphofructokinase-1.
Nat Commun, 15, 2024
|
|
5O86
 
 | |
7W6D
 
 | |
6S7Z
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-((3,4-Dihydroisoquinolin-2(1H)-yl)sulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
3ZCR
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4-tert- butyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2.07 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
1BXS
 
 | | SHEEP LIVER CLASS 1 ALDEHYDE DEHYDROGENASE WITH NAD BOUND | | Descriptor: | ALDEHYDE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moore, S.A, Baker, H.M, Blythe, T.J, Kitson, K.E, Kitson, T.M, Baker, E.N. | | Deposit date: | 1998-10-08 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases.
Structure, 6, 1998
|
|
7N6B
 
 | | Structure of MmpL3 reconstituted into lipid nanodisc in the TMM bound state | | Descriptor: | 6-O-[(2S)-2-{(1S)-18-[(1R,2R)-2-hexylcyclopropyl]-1-hydroxyoctadecyl}tricosanoyl]-alpha-D-glucopyranosyl alpha-D-glucopyranoside, MmpL3 transporter | | Authors: | Su, C.C, Yu, E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
4ZNZ
 
 | | Crystal structure of Escherichia coli carbonic anhydrase (YadF) in complex with Zn - artifact of purification | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Gasiorowska, O.A, Niedzialkowska, E, Porebski, P.J, Handing, K.B, Shabalin, I.G, Cymborowski, M.T, Minor, W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
6RRO
 
 | |
5NTN
 
 | | Structural states of RORgt: X-ray elucidation of molecular mechanisms and binding interactions for natural and synthetic compounds | | Descriptor: | (5R,10S,13R,14R,17R)-17-((R,E)-7-hydroxy-6-methylhept-5-en-2-yl)-4,4,10,13,14-pentamethyl-1,2,5,6,10,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthrene-3,7(4H)-dione, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Kallen, J. | | Deposit date: | 2017-04-28 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural States of ROR gamma t: X-ray Elucidation of Molecular Mechanisms and Binding Interactions for Natural and Synthetic Compounds.
ChemMedChem, 12, 2017
|
|
3ZCQ
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4- trifluoromethyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2. 15 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-{[4-(trifluoromethyl)benzoyl]carbamoyl}-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
1G05
 
 | | HETEROCYCLE-BASED MMP INHIBITOR WITH P2'SUBSTITUENTS | | Descriptor: | 1-BENZYL-3-(4-METHOXY-BENZENESULFONYL)-6-OXO-HEXAHYDRO-PYRIMIDINE-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Pikul, S, Dunham, K.M, Almstead, N.G, De, B, Natchus, M.G, Taiwo, Y.O, Williams, L.E, Hynd, B.A, Hsieh, L.C, Janusz, M.J. | | Deposit date: | 2000-10-05 | | Release date: | 2001-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Heterocycle-based MMP inhibitors with P2' substituents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
2V4N
 
 | | Crystal structure of Salmonella typhimurium SurE at 1.7 angstrom resolution in orthorhombic form | | Descriptor: | GLYCEROL, MAGNESIUM ION, MULTIFUNCTIONAL PROTEIN SUR E, ... | | Authors: | Anju, P, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-09-26 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies on a Mesophilic Stationary Phase Survival Protein (Sur E) from Salmonella Typhimurium
FEBS J., 275, 2008
|
|
