5AN3
 
 | | Structure of an Sgt1-Skp1 Complex | | Descriptor: | SGT1, SUPPRESSOR OF KINETOCHORE PROTEIN 1 | | Authors: | Willhoft, O, Vaughan, C.K. | | Deposit date: | 2015-09-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The crystal structure of the Sgt1-Skp1 complex: the link between Hsp90 and both SCF E3 ubiquitin ligases and kinetochores.
Sci Rep, 7, 2017
|
|
7W8A
 
 | | Babesia orientalis lactate dehydrogenase, BoLDH apo | | Descriptor: | lactate dehydrogenase | | Authors: | Yu, L. | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | The Structural Basis of Babesia orientalis Lactate Dehydrogenase.
Front Cell Infect Microbiol, 11, 2021
|
|
4X9S
 
 | | CRYSTAL STRUCTURE OF HISAP FROM STREPTOMYCES SP. MG1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
5ANU
 
 | | MTH1 in complex with compound 15 | | Descriptor: | 13-(METHYLAMINO)-23,24,25-TRIOXA-17,18,19,21-TETRAZATETRACYCLO-TRICOSA-1(3),2(10),4(11),12(14),13(18),16(19)-HEXAN-15-ONE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, DIMETHYL SULFOXIDE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5ANW
 
 | | MTH1 in complex with compound 24 | | Descriptor: | 2-[4-(2-AMINOQUINAZOLIN-4-YL)PHENYL]-N,N-DIMETHYL-ACETAMIDE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
7W1G
 
 | | Crystal structure of YfiH with C107A mutation in complex with UDP-MurNAc-L-Serine | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, (2S)-2-[[(2R)-2-[(2R,3R,4R,5S,6R)-3-acetamido-2-[[[(2R,3S,4R,5R)-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-6-(hydroxymethyl)-5-oxidanyl-oxan-4-yl]oxypropanoyl]amino]-3-oxidanyl-propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lee, S.H, Hsieh, K.Y, Lee, M.S, Chang, C.I. | | Deposit date: | 2021-11-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for the Peptidoglycan-Editing Activity of YfiH.
Mbio, 13, 2021
|
|
4X9Q
 
 | | MnSOD-3 Room Temperature Structure | | Descriptor: | MALONATE ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Hunter, T, Bonetta, R, Stewart, E.E. | | Deposit date: | 2014-12-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structure of the Caenorhabditis elegans manganese superoxide dismutase MnSOD-3-azide complex.
Protein Sci., 24, 2015
|
|
1Z11
 
 | | Crystal Structure of Human Microsomal P450 2A6 with Methoxsalen Bound | | Descriptor: | METHOXSALEN, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450, ... | | Authors: | Yano, J.K, Hsu, M.H, Griffin, K.J, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-03-02 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen
Nat.Struct.Mol.Biol., 12, 2005
|
|
5BX3
 
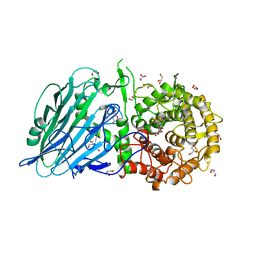 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, CALCIUM ION, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tankrathok, A, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
5VOY
 
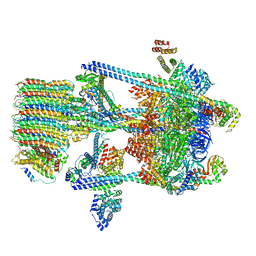 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 2) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5C1I
 
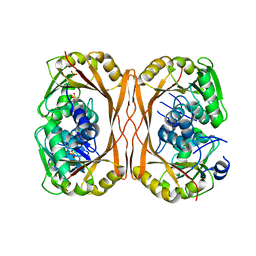 | | m1A58 tRNA methyltransferase mutant - D170A | | Descriptor: | SULFATE ION, tRNA (adenine(58)-N(1))-methyltransferase TrmI | | Authors: | Ponchon, L, Degut, C, Folly-Klan, M, Barraud, P, Tisne, C. | | Deposit date: | 2015-06-14 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The m1A58 modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.
Biophys.Chem., 210, 2016
|
|
7WBE
 
 | |
7WBF
 
 | | Crystal structure of lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Nam, K.H. | | Deposit date: | 2021-12-16 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Processing of Multicrystal Diffraction Patterns in Macromolecular Crystallography Using Serial Crystallography Programs.
Crystals, 12, 2022
|
|
7WBD
 
 | |
5E2U
 
 | |
5E5W
 
 | | Hemagglutinin-esterase-fusion mutant structure of influenza D virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
4XL1
 
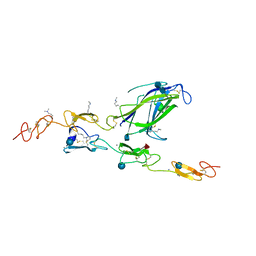 | | Complex of Notch1 (EGF11-13) bound to Delta-like 4 (N-EGF1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural biology. Structural basis for Notch1 engagement of Delta-like 4.
Science, 347, 2015
|
|
5E78
 
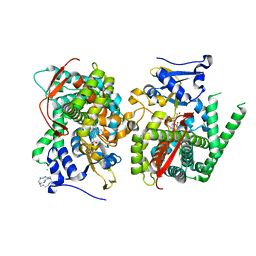 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | Descriptor: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
5E0C
 
 | |
4XLW
 
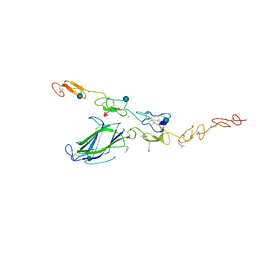 | | Complex of Notch1 (EGF11-13) bound to Delta-like 4 (N-EGF2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Delta-like protein, ... | | Authors: | Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Structural biology. Structural basis for Notch1 engagement of Delta-like 4.
Science, 347, 2015
|
|
4C6F
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 6.5 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, DIHYDROOROTASE, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
5DKX
 
 | | Crystal structure of glucosidase II alpha subunit (Tris-bound from) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CHLORIDE ION | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
7WH1
 
 | | structure of C elegans BCMO-2 | | Descriptor: | Beta-Carotene 15,15'-MonoOxygenase, FE (III) ION, GLYCEROL, ... | | Authors: | Pan, W, Liu, L. | | Deposit date: | 2021-12-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of Nonheme Iron Enzymes BCMO-1 and BCMO-2 from Caenorhabditis elegans .
Front Mol Biosci, 9, 2022
|
|
5DM9
 
 | |
5E3H
 
 | | Structural Basis for RNA Recognition and Activation of RIG-I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Jiang, F, Miller, M.T, Marcotrigiano, J. | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of RNA recognition and activation by innate immune receptor RIG-I.
Nature, 479, 2011
|
|
