8V5C
 
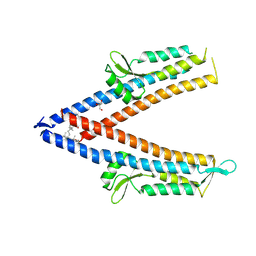 | | IpaD (122-321) Bound to Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCEROL, Invasin IpaD | | Authors: | Barker, S.A, Dickenson, N.E, Johnson, S.J. | | Deposit date: | 2023-11-30 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of the IpaD pi-helix reveals critical roles in DOC interaction, T3SS apparatus maturation, and Shigella virulence.
J.Biol.Chem., 300, 2024
|
|
6Q4C
 
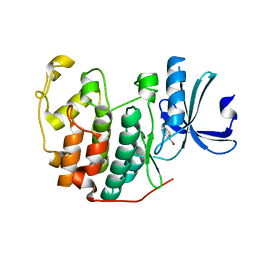 | | CDK2 in complex with FragLite16 | | Descriptor: | 4-bromanyl-1,8-naphthyridine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4F
 
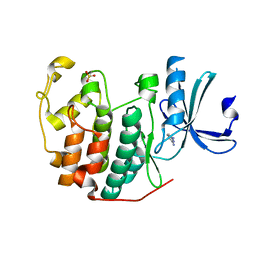 | | CDK2 in complex with FragLite32 | | Descriptor: | Cyclin-dependent kinase 2, PYRIDINE-2,6-DIAMINE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
5JCS
 
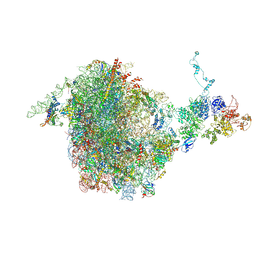 | | CRYO-EM STRUCTURE OF THE RIX1-REA1 PRE-60S PARTICLE | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Barrio-Garcia, C, Thoms, M, Flemming, D, Kater, L, Berninghausen, O, Bassler, J, Beckmann, R, Hurt, E. | | Deposit date: | 2016-04-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling
Nat.Struct.Mol.Biol., 23, 2016
|
|
5NGJ
 
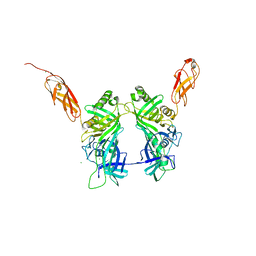 | | Crystal structure of pb6, major tail tube protein of bacteriophage T5 | | Descriptor: | CHLORIDE ION, Tail tube protein | | Authors: | Arnaud, C.-A, Effantin, G, Vives, C, Engilberge, S, Bacia, M, Boulanger, P, Girard, E, Schoehn, G, Breyton, C. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection.
Nat Commun, 8, 2017
|
|
5JIL
 
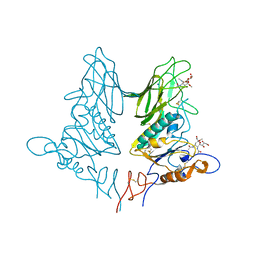 | | Crystal structure of rat coronavirus strain New-Jersey Hemagglutinin-Esterase in complex with 4N-acetyl sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1LUV
 
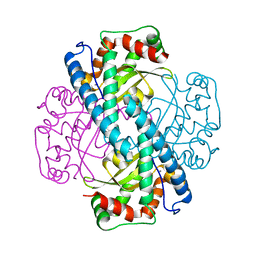 | | CATALYTIC AND STRUCTURAL EFFECTS OF AMINO-ACID SUBSTITUTION AT HIS 30 IN HUMAN MANGANESE SUPEROXIDE DISMUTASE: INSERTION OF VAL CGAMMA INTO THE SUBSTRATE ACCESS CHANNEL | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn] | | Authors: | Hearn, A.S, Stroupe, M.E, Ramilo, C.A, Luba, J.P, Cabelli, D.E, Tainer, J.A, Silverman, D.N. | | Deposit date: | 2002-05-23 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalytic and structural effects of amino acid substitution at histidine 30 in human manganese superoxide dismutase: insertion of valine C gamma into the substrate access channel
Biochemistry, 42, 2003
|
|
6Q96
 
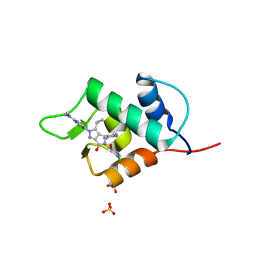 | |
6GQM
 
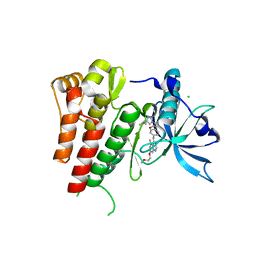 | | Crystal structure of human c-KIT kinase domain in complex with a small molecule inhibitor, AZD3229 | | Descriptor: | CHLORIDE ION, Mast/stem cell growth factor receptor Kit, ~{N}-[4-[[5-fluoranyl-7-(2-methoxyethoxy)quinazolin-4-yl]amino]phenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
7VGN
 
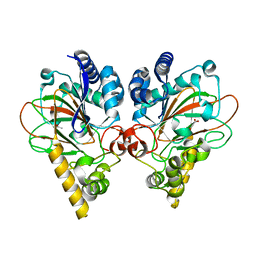 | | Crystal structure of CmnC | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CmnC, ... | | Authors: | Huang, S.J, Hsiao, Y.H, Lin, E.C, Lee, Y.C, Zheng, Y.Z, Chang, C.Y. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
5JMO
 
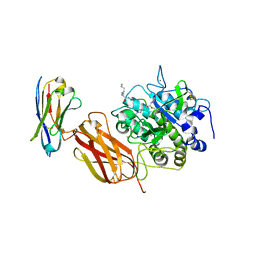 | |
5KVA
 
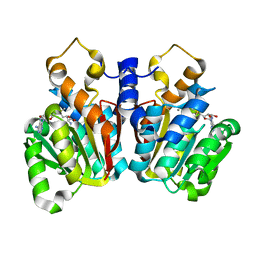 | | Crystal Structure of sorghum caffeoyl-CoA O-methyltransferase (CCoAOMT) | | Descriptor: | CALCIUM ION, S-ADENOSYLMETHIONINE, caffeoyl-CoA O-methyltransferase | | Authors: | Walker, A.M, Sattler, S.A, Regner, M, Jones, J.P, Ralph, J, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | The Structure and Catalytic Mechanism of Sorghum bicolor Caffeoyl-CoA O-Methyltransferase.
Plant Physiol., 172, 2016
|
|
7VGL
 
 | | Crystal structure of CmnC | | Descriptor: | ACETATE ION, CmnC | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Zheng, Y.Z, Chang, C.Y. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
2VC7
 
 | | Structural basis for natural lactonase and promiscuous phosphotriesterase activities | | Descriptor: | (4S)-4-(decanoylamino)-5-hydroxy-3,4-dihydro-2H-thiophenium, 1,2-ETHANEDIOL, ARYLDIALKYLPHOSPHATASE, ... | | Authors: | Elias, M, Dupuy, J, Merone, L, Mandrich, L, Moniot, S, Rochu, D, Lecomte, C, Rossi, M, Masson, P, Manco, G, Chabriere, E. | | Deposit date: | 2007-09-19 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Natural Lactonase and Promiscuous Phosphotriesterase Activities.
J.Mol.Biol., 379, 2008
|
|
6PS5
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to Propranolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
8UOA
 
 | | Structure of the synaptic vesicle protein 2A Luminal domain in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, Synaptic vesicle glycoprotein 2A, ... | | Authors: | Mittal, A, Martin, M.F, Levin, E, Adams, C, Yang, M, Ledecq, M, Horanyi, P.S, Coleman, J.A. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of synaptic vesicle protein 2A and 2B bound to anticonvulsants.
Nat.Struct.Mol.Biol., 2024
|
|
6TTJ
 
 | | Neutral invertase 2 from Arabidopsis thaliana | | Descriptor: | Alkaline/neutral invertase CINV1 | | Authors: | Tsirkone, V.G, Osipov, E.M, Beelen, S, Strelkov, S.V. | | Deposit date: | 2019-12-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.392 Å) | | Cite: | Crystal structure of Arabidopsis thaliana neutral invertase 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6GUE
 
 | | CDK2/CyclinA in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
4CBC
 
 | |
6GUR
 
 | | Siderophore hydrolase EstB from Aspergillus fumigatus in complex with TAFC | | Descriptor: | (~{Z})-5-[(1~{S},2~{S})-2-acetamido-1-oxidanyl-5-[oxidanyl(propanoyl)amino]pentoxy]-~{N},3-dimethyl-~{N}-oxidanyl-pent-2-enamide, CARBONATE ION, FE (III) ION, ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7VF8
 
 | | Crystal Structure of HasAp with Co-5-octaethyloxaporphyrinium cation | | Descriptor: | CITRIC ACID, Co-5-octaethyloxaporphyrinium cation, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Takiguchi, A, Sakakibara, E, Sugimoto, H, Shoji, O, Shinokubo, H. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of HasAp with Co-5-octaethyloxaporphyrinium cation
To Be Published
|
|
1ZJF
 
 | | 12mer-spd-P4N | | Descriptor: | 5'-D(*AP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*C)-3' | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
1K4J
 
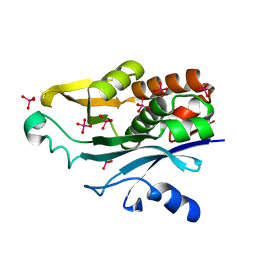 | | Crystal Structure of the Acyl-homoserinelactone Synthase EsaI Complexed with Rhenate | | Descriptor: | PERRHENATE, acyl-homoserinelactone synthase EsaI | | Authors: | Watson, W.T, Minogue, T.D, Val, D.L, Beck von Bodman, S, Churchill, M.E.A. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing.
Mol.Cell, 9, 2002
|
|
7LQH
 
 | | Cryo-EM of KFE8 thinner nanotube (class 2, 2-sub-1) | | Descriptor: | KFE8 peptide | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Deterministic chaos in the self-assembly of beta sheet nanotubes from an amphipathic oligopeptide.
Matter, 4, 2021
|
|
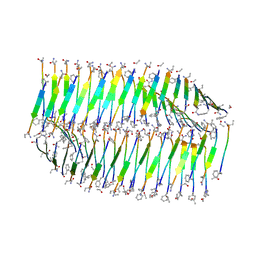
7LQF
 
 | |
