3JXC
 
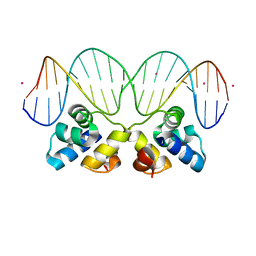 | |
3K0S
 
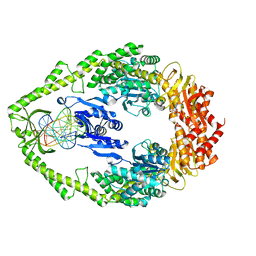 | | Crystal structure of E.coli DNA mismatch repair protein MutS, D693N mutant, in complex with GT mismatched DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*C*AP*CP*T*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Reumer, G.A, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2009-09-25 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS.
J.Biol.Chem., 285, 2010
|
|
3K2K
 
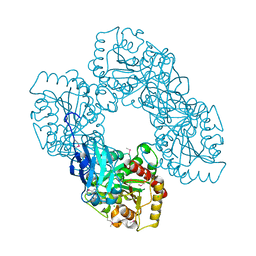 | |
3K2R
 
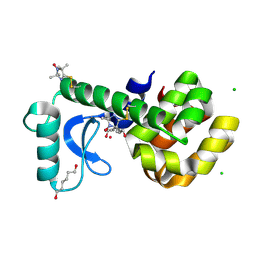 | | Crystal Structure of Spin Labeled T4 Lysozyme Mutant K65V1/R76V1 | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, Lysozyme, ... | | Authors: | Toledo Warshaviak, D, Cascio, D, Khramtsov, V.V, Hubbell, W.L. | | Deposit date: | 2009-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Spin Labeled T4 Lysozyme Mutant K65V1/R76V1
To be Published
|
|
3K2X
 
 | |
3K3W
 
 | | Thermostable Penicillin G acylase from Alcaligenes faecalis in orthorhombic form | | Descriptor: | CALCIUM ION, Penicillin G acylase | | Authors: | Varshney, N.K, Kumar, R.S, Ignatova, Z, Dodson, E, Suresh, C.G. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystallization and X-ray structure analysis of a thermostable penicillin G acylase from Alcaligenes faecalis.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3K5C
 
 | | Human BACE-1 complex with NB-216 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({1-[3-(1-methylethyl)phenyl]cyclopropyl}amino)ethyl]-19-(methoxymethyl)-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Macrocyclic BACE-1 inhibitors acutely reduce Abeta in brain after po application.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3K69
 
 | |
3K6V
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Citrate-Bound Open Form | | Descriptor: | CITRIC ACID, Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3JTJ
 
 | | 3-deoxy-manno-octulosonate cytidylyltransferase from Yersinia pestis | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase, IMIDAZOLE | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-12 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | X-ray crystal structure of 3-deoxy-manno-octulosonate cytidylyltransferase from Yersinia pestis.
To be Published
|
|
3JTR
 
 | | Mutations in Cephalosporin Acylase Affecting Stability and Autoproteolysis | | Descriptor: | GLYCEROL, Glutaryl 7-aminocephalosporanic acid acylase | | Authors: | Cho, K.J, Kim, J.K, Lee, J.H, Shin, H.J, Park, S.S, Kim, K.H. | | Deposit date: | 2009-09-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural features of cephalosporin acylase reveal the basis of autocatalytic activation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3JUY
 
 | |
3JWJ
 
 | |
3JY9
 
 | | Janus Kinase 2 Inhibitors | | Descriptor: | (3S)-3-(4-hydroxyphenyl)-1,5-dihydro-1,5,12-triazabenzo[4,5]cycloocta[1,2,3-cd]inden-4(3H)-one, SODIUM ION, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Ledeboer, M.W, Pierce, A.C. | | Deposit date: | 2009-09-21 | | Release date: | 2009-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Janus kinase 2 inhibitors. Synthesis and characterization of a novel polycyclic azaindole.
J.Med.Chem., 52, 2009
|
|
3JZ6
 
 | | Crystal structure of Mycobacterium smegmatis Branched Chain Aminotransferase in complex with pyridoxal-5'-phosphate at 1.9 angstrom. | | Descriptor: | Branched-chain amino acid aminotransferase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Castell, A, Mille, C, Unge, T. | | Deposit date: | 2009-09-23 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of mycobacterial branched-chain aminotransferase: implications for inhibitor design.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JZV
 
 | |
3K03
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-DTPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K29
 
 | | Structure of a putative YscO homolog CT670 from Chlamydia trachomatis | | Descriptor: | Putative uncharacterized protein | | Authors: | Lam, R, Singer, A, Skarina, T, Onopriyenko, O, Bochkarev, A, Brunzelle, J.S, Edwards, A.M, Anderson, W.F, Chirgadze, N.Y, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and protein-protein interaction studies on Chlamydia trachomatis protein CT670 (YscO Homolog).
J.Bacteriol., 192, 2010
|
|
3K2L
 
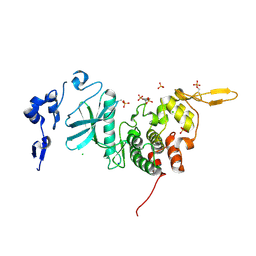 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) | | Descriptor: | CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2, SODIUM ION, ... | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Soundararajan, M, Krojer, T, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of Down Syndrome Kinases, DYRKs, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
3K3P
 
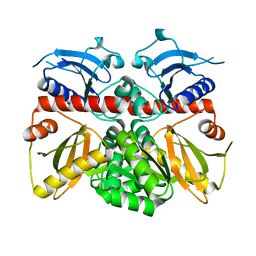 | |
3JQ7
 
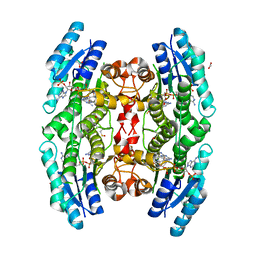 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 6-phenylpteridine-2,4,7-triamine (DX2) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-phenylpteridine-2,4,7-triamine, ACETATE ION, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQF
 
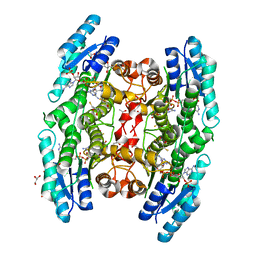 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 1,3,5-triazine-2,4,6-triamine (AX2) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1,3,5-triazine-2,4,6-triamine, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3K5U
 
 | | Identification, SAR Studies and X-ray Cocrystal Analysis of a Novel Furano-pyrimidine Aurora Kinase A Inhibitor | | Descriptor: | 2-[(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)AMINO]ETHANOL, Serine/threonine-protein kinase 6 | | Authors: | Wu, J.S, Leou, J.S, Coumar, M.S, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2009-10-08 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification, SAR studies, and X-ray co-crystallographic analysis of a novel furanopyrimidine aurora kinase A inhibitor
Chemmedchem, 5, 2010
|
|
3JRN
 
 | | Crystal structure of TIR domain from Arabidopsis Thaliana | | Descriptor: | ARSENIC, AT1G72930 protein | | Authors: | Chan, S.L, Mukasa, T, Santelli, E, Low, L.Y, Pascual, J. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants.
Protein Sci., 19, 2009
|
|
3JRT
 
 | | Structure from the mobile metagenome of V. paracholerae: Integron cassette protein Vpc_cass2 | | Descriptor: | Integron cassette protein Vpc_cass2 | | Authors: | Harrop, S.J, Deshpande, C, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Cuff, M, Edwards, A, Savchenko, A, Joachimiak, A, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-22 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites.
Plos One, 8, 2013
|
|
