3F2M
 
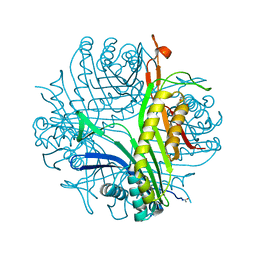 | | Urate oxidase complexed with 8-azaxanthine at 150 MPa | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Girard, E, Kahn, R, Fourme, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function perturbation and dissociation of tetrameric urate oxidase by high hydrostatic pressure.
Biophys.J., 98, 2010
|
|
3F5Q
 
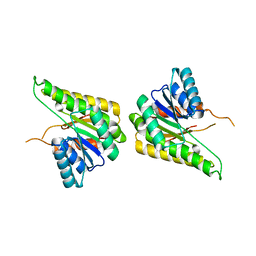 | | CRYSTAL STRUCTURE OF putative short chain dehydrogenase from Escherichia coli CFT073 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
3FNA
 
 | |
3F62
 
 | |
3F6B
 
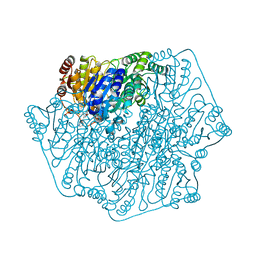 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor PAA | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
3FOA
 
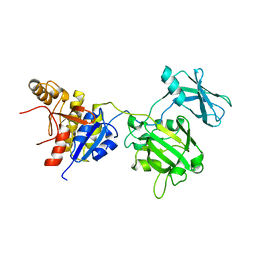 | | Crystal structure of the bacteriophage T4 tail sheath protein, deletion mutant gp18M | | Descriptor: | Tail sheath protein Gp18 | | Authors: | Aksyuk, A.A, Leiman, P.G, Kurochkina, L.P, Shneider, M.M, Kostyuchenko, V.A, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2008-12-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria.
Embo J., 28, 2009
|
|
3F75
 
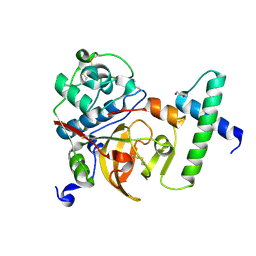 | | Activated Toxoplasma gondii cathepsin L (TgCPL) in complex with its propeptide | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2008-11-07 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Toxoplasma gondii cathepsin L is the primary target of the invasion-inhibitory compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl.
J.Biol.Chem., 284, 2009
|
|
3F9E
 
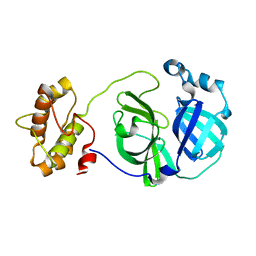 | |
3EYG
 
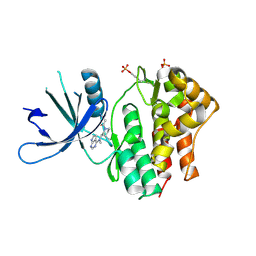 | | Crystal structures of JAK1 and JAK2 inhibitor complexes | | Descriptor: | 3-{(3R,4R)-4-methyl-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}-3-oxopropanenitrile, Tyrosine-protein kinase | | Authors: | Williams, N.K, Bamert, R.S, Patell, O, Wang, C, Walden, P.M, Fantino, E. | | Deposit date: | 2008-10-20 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains.
J.Mol.Biol., 387, 2009
|
|
3FAZ
 
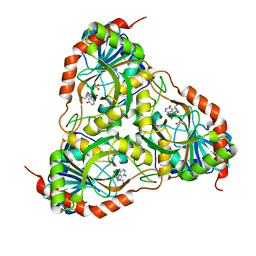 | | Crystal structure of Schistosoma mansoni purine nucleoside phosphorylase in complex with inosine | | Descriptor: | INOSINE, Purine-nucleoside phosphorylase, SULFATE ION | | Authors: | Pereira, H.M, Garratt, R.C, Oliva, G. | | Deposit date: | 2008-11-18 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3FCR
 
 | |
3FD0
 
 | |
3FDC
 
 | | Crystal Structure of Avidin | | Descriptor: | Avidin, iron(II) tetracyano-5-(2-Oxo-hexahydro-thieno[3,4-d]imidazol-6-yl)-pentanoic acid (4'-methyl-[2,2']bipyridinyl-4-ylmethyl)-amide | | Authors: | Barker, K.D, Sazinsky, M.H, Eckermann, A.L, Abajian, C, Hartings, M.R, Rosenzweig, A.C, Meade, T.J. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Binding and the Electronic Properties of Iron(II) Complexes: An Electrochemical and Optical Investigation of Outer Sphere Effects.
Bioconjug.Chem., 20, 2009
|
|
3FDH
 
 | |
3F78
 
 | | Crystal structure of wild type LFA1 I domain complexed with isoflurane | | Descriptor: | 1-CHLORO-2,2,2-TRIFLUOROETHYL DIFLUOROMETHYL ETHER, GLYCEROL, Integrin alpha-L, ... | | Authors: | Zhang, H, Wang, J.-H. | | Deposit date: | 2008-11-07 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of isoflurane bound to integrin LFA-1 supports a unified mechanism of volatile anesthetic action in the immune and central nervous systems.
Faseb J., 23, 2009
|
|
3FDU
 
 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Acinetobacter baumannii | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase/isomerase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Acinetobacter baumannii
To be Published
|
|
3FH6
 
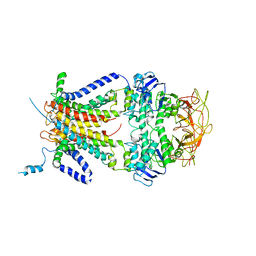 | | Crystal structure of the resting state maltose transporter from E. coli | | Descriptor: | Maltose transport system permease protein malF, Maltose transport system permease protein malG, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Khare, D, Oldham, M.L, Orelle, C, Davidson, A.L, Chen, J. | | Deposit date: | 2008-12-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Alternating access in maltose transporter mediated by rigid-body rotations.
Mol.Cell, 33, 2009
|
|
3FHT
 
 | |
3FJX
 
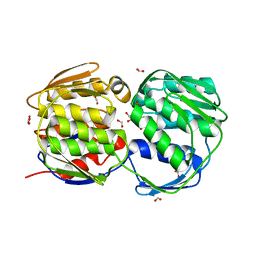 | | E. coli EPSP synthase (T97I) liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3FLE
 
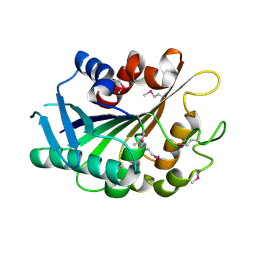 | | SE_1780 protein of unknown function from Staphylococcus epidermidis. | | Descriptor: | SE_1780 protein | | Authors: | Osipiuk, J, Hatzos, C, Clancy, S, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | X-ray crystal structure of SE_1780 protein of unknown function from Staphylococcus epidermidis.
To be Published
|
|
3FLP
 
 | |
3FMZ
 
 | |
3FHX
 
 | | Crystal structure of D235A mutant of human pyridoxal kinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Safo, M.K, Gandhi, A.K, Musayev, F.N, Ghatge, M, Di Salvo, M.L, Schirch, V. | | Deposit date: | 2008-12-10 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetic and structural studies of the role of the active site residue Asp235 of human pyridoxal kinase.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3FON
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFEAI | | Descriptor: | Beta-2-microglobulin, MHC, Peptide | | Authors: | Malashkevich, V.N, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Brims, D.R, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
3FP5
 
 | | Crystal structure of ACBP from Moniliophthora perniciosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acyl-CoA Binding Protein, ZINC ION | | Authors: | Monzani, P.S, Pereira, H.M, Melo, F.A, Meirelles, F.V, Oliva, G, Cascardo, J.C.M. | | Deposit date: | 2009-01-04 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A new topology of ACBP from Moniliophthora perniciosa.
Biochim.Biophys.Acta, 1804, 2010
|
|
