4M2Y
 
 | | Structure of human DNA polymerase beta complexed with 8-BrG as the template base in a 1-nucleotide gapped DNA | | Descriptor: | DNA polymerase beta, SODIUM ION, down-primer, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-05 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
1EN4
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Q146H MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1EQP
 
 | |
3VKE
 
 | |
7NPQ
 
 | |
1EQX
 
 | | SOLUTION STRUCTURE DETERMINATION AND MUTATIONAL ANALYSIS OF THE PAPILLOMAVIRUS E6-INTERACTING PEPTIDE OF E6AP | | Descriptor: | PAPILLOMAVIRUS E6-ASSOCIATED PROTEIN | | Authors: | Be, X, Hong, Y, Androphy, E.J, Chen, J.J, Baleja, J.D. | | Deposit date: | 2000-04-06 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure determination and mutational analysis of the papillomavirus E6 interacting peptide of E6AP.
Biochemistry, 40, 2001
|
|
1ENT
 
 | |
1ERE
 
 | |
1ET0
 
 | | CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Nakai, T, Mizutani, H, Miyahara, I, Hirotsu, K, Takeda, S, Jhee, K.H, Yoshimura, T, Esaki, N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of 4-amino-4-deoxychorismate lyase from Escherichia coli.
J.Biochem.(Tokyo), 128, 2000
|
|
1EO6
 
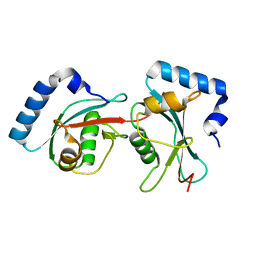 | | CRYSTAL STRUCTURE OF GATE-16 | | Descriptor: | GOLGI-ASSOCIATED ATPASE ENHANCER OF 16 KD | | Authors: | Paz, Y, Elazar, Z, Fass, D. | | Deposit date: | 2000-03-22 | | Release date: | 2000-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p.
J.Biol.Chem., 275, 2000
|
|
1EOD
 
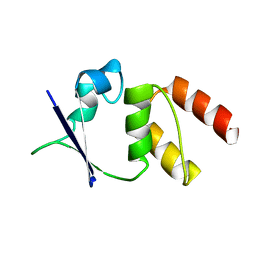 | | CRYSTAL STRUCTURE OF THE N136D MUTANT OF A SHAKER T1 DOMAIN | | Descriptor: | POTASSIUM CHANNEL KV1.1 | | Authors: | Nanao, M.H, Cushman, S.J, Jahng, A.W, DeRubeis, D, Choe, S, Pfaffinger, P.J. | | Deposit date: | 2000-03-22 | | Release date: | 2000-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Voltage dependent activation of potassium channels is coupled to T1 domain structure.
Nat.Struct.Biol., 7, 2000
|
|
1ET8
 
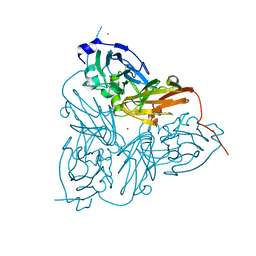 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASN MUTANT FROM ALCALIGENES FAECALIS | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
1EOI
 
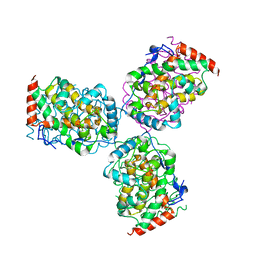 | | CRYSTAL STRUCTURE OF ACID PHOSPHATASE FROM ESCHERICHIA BLATTAE COMPLEXED WITH THE TRANSITION STATE ANALOG MOLYBDATE | | Descriptor: | ACID PHOSPHATASE, MOLYBDATE ION | | Authors: | Ishikawa, K, Mihara, Y, Gondoh, K, Suzuki, E, Asano, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2001-03-23 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate.
EMBO J., 19, 2000
|
|
1EUE
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oganesyan, V, Zhang, X. | | Deposit date: | 2000-04-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of redox potential in electron transfer proteins: effects of complex formation on the active site microenvironment of cytochrome b5.
FARADAY DISC.CHEM.SOC, 116, 2001
|
|
1EP5
 
 | |
1EUO
 
 | | Crystal structure of nitrophorin 2 (prolixin-S) | | Descriptor: | AMMONIA, NITROPHORIN 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Andersen, J.F, Montfort, W.R. | | Deposit date: | 2000-04-17 | | Release date: | 2000-05-10 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of nitrophorin 2. A trifunctional antihemostatic protein from the saliva of Rhodnius prolixus
J.Biol.Chem., 275, 2000
|
|
3VNF
 
 | | Structure of the ebolavirus protein VP24 from Sudan | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Zhang, A.P.P. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold
Plos Pathog., 8, 2012
|
|
1E9D
 
 | | Mutant human thymidylate kinase (F105Y) complexed with AZTMP and ADP | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N, Lavie, A, Padiyar, S, Brundiers, R, Veit, T, Reintein, J, Goody, R.S, Konrad, M, Schlichting, I. | | Deposit date: | 2000-10-10 | | Release date: | 2001-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Potentiating Azt Activation: Structures of Wildtype and Mutant Human Thymidylate Kinase Suggest Reasons for the Mutants' Improved Kinetics with the HIV Prodrug Metabolite Aztmp
J.Mol.Biol., 304, 2000
|
|
1EPV
 
 | | ALANINE RACEMASE WITH BOUND INHIBITOR DERIVED FROM D-CYCLOSERINE | | Descriptor: | ALANINE RACEMASE, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Fenn, T.D, Stamper, G.F, Morollo, A.A, Ringe, D. | | Deposit date: | 2000-03-29 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A side reaction of alanine racemase: transamination of cycloserine.
Biochemistry, 42, 2003
|
|
1ETS
 
 | |
1EQH
 
 | | THE 2.7 ANGSTROM MODEL OF OVINE COX-1 COMPLEXED WITH FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, PROSTAGLANDIN H2 SYNTHASE-1, ... | | Authors: | Loll, P.J, Selinsky, B.S, Gupta, K, Sharkey, C.T. | | Deposit date: | 2000-04-04 | | Release date: | 2001-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations.
Biochemistry, 40, 2001
|
|
1ERB
 
 | |
1ES7
 
 | |
1ESG
 
 | |
1ESX
 
 | | 1H, 15N AND 13C STRUCTURE OF THE HIV-1 REGULATORY PROTEIN VPR : COMPARISON WITH THE N-AND C-TERMINAL DOMAIN STRUCTURE, (1-51)VPR AND (52-96)VPR | | Descriptor: | VPR PROTEIN | | Authors: | Wecker, K, Morellet, N, Bouaziz, S, Roques, B. | | Deposit date: | 2000-04-11 | | Release date: | 2001-04-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HIV-1 regulatory protein Vpr in H2O/trifluoroethanol. Comparison with the Vpr N-terminal (1-51) and C-terminal (52-96) domains.
Eur.J.Biochem., 269, 2002
|
|
