6MT8
 
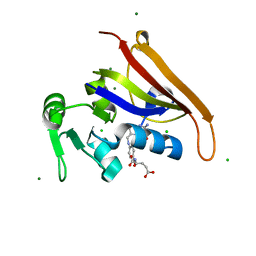 | | E. coli DHFR complex modeled with two ligand states | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, DIHYDROFOLIC ACID, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Time-resolved x-ray crystallography capture of a slow reaction tetrahydrofolate intermediate.
Struct Dyn., 6, 2019
|
|
6N04
 
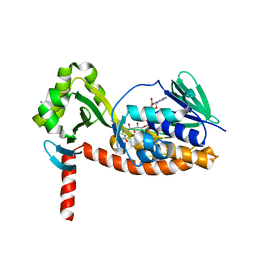 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
6N3U
 
 | |
6MTH
 
 | | E. coli DHFR complex modeled with three ligand states | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, DIHYDROFOLIC ACID, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Time-resolved x-ray crystallography capture of a slow reaction tetrahydrofolate intermediate.
Struct Dyn., 6, 2019
|
|
6MTM
 
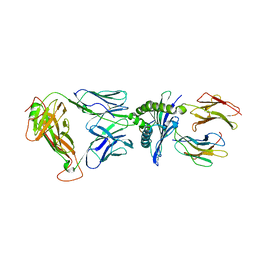 | | Crystal Structure of EM2 TCR in complex with HLA-B*37:01-NP338 | | Descriptor: | Beta-2-microglobulin, EM2 TCR alpha chain, EM2 TCR beta chain, ... | | Authors: | Gras, S. | | Deposit date: | 2018-10-19 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Broad CD8+T cell cross-recognition of distinct influenza A strains in humans.
Nat Commun, 9, 2018
|
|
6MZI
 
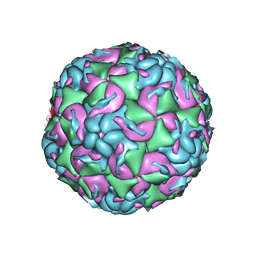 | | CryoEM structure of human enterovirus D68 expanded 1 particle (pH 6.5, 4 degrees Celsius, 3 min) | | Descriptor: | viral protein 1, viral protein 2, viral protein 3, ... | | Authors: | Liu, Y, Rossmann, M.G. | | Deposit date: | 2018-11-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Molecular basis for the acid-initiated uncoating of human enterovirus D68.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N0W
 
 | |
6N4J
 
 | |
6N2J
 
 | | Tetrahydropyridopyrimidines as Covalent Inhibitors of KRAS-G12C | | Descriptor: | 1-{4-[7-(naphthalen-1-yl)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vigers, G.P. | | Deposit date: | 2018-11-13 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Tetrahydropyridopyrimidines as Irreversible Covalent Inhibitors of KRAS-G12C with In Vivo Activity.
ACS Med Chem Lett, 9, 2018
|
|
6N4K
 
 | |
6N3F
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to SF3b1 ULM5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Leach, J.R, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
6WY5
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-37 A.K.A 7-(1-phenyl-3-(((1S,3S)-3-phenyl-2,3-dihydro-1H-inden-1-yl)amino)propyl)-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1-phenyl-3-{[(1S,3S)-3-phenyl-2,3-dihydro-1H-inden-1-yl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6N9E
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic CC-Pmn and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
4WJG
 
 | | Structure of T. brucei haptoglobin-hemoglobin receptor binding to human haptoglobin-hemoglobin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin, ... | | Authors: | Stoedkilde, K, Torvund-Jensen, M, Moestrup, S.K, Andersen, C.B.F. | | Deposit date: | 2014-09-30 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for trypanosomal haem acquisition and susceptibility to the host innate immune system.
Nat Commun, 5, 2014
|
|
6NVD
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with triazole linked compound 9 | | Descriptor: | 2',5'-dideoxy-5'-[4-({[(1S,2R)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetyl}amino)-1H-1,2,3-triazol-1-yl]cytidine, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with triazole linked compound 9
To Be Published
|
|
6NKA
 
 | |
6WY7
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-41 A.K.A 7-[1-phenyl-3-({4-phenylbicyclo[2.2.2]octan-1-yl}amino)propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{(1R)-1-phenyl-3-[(4-phenylbicyclo[2.2.2]octan-1-yl)amino]propyl}-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6WY0
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-40 A.K.A 7-[(1R)-1-phenyl-3-{[(1r,4r)-4-phenylcyclohexyl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{(1R)-1-phenyl-3-[(trans-4-phenylcyclohexyl)amino]propyl}-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6NR6
 
 | | Crystal Structure of Staphylococcus pseudintermedius SbnI in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional transcriptional regulator/O-phospho-L-serine synthase SbnI, MAGNESIUM ION | | Authors: | Verstraete, M.M, Morales, L.D, Kobylarz, M.J, Loutet, S.A, Laakso, H.A, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2019-01-22 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The heme-sensitive regulator SbnI has a bifunctional role in staphyloferrin B production by Staphylococcus aureus .
J.Biol.Chem., 294, 2019
|
|
6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6WXZ
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-29 A.K.A 7-(1,2-DIPHENYLETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRIDIN-5-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1,2-diphenylethyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6NLI
 
 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 11) | | Descriptor: | 4-{[(2-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NOT
 
 | |
6NLM
 
 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 15) | | Descriptor: | 4-{[3-(4-hydroxy-2-methoxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NNC
 
 | | Structure of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
