3V6M
 
 | | Inhibition of caspase-6 activity by single mutation outside the active site | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
3UNS
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4FUV
 
 | | Crystal Structure of Acinetobacter baumannii CarO | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SULFATE ION, porin protein associated with imipenem resistance | | Authors: | Eren, E, van den Berg, B. | | Deposit date: | 2012-06-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Outer membrane transport of amino acids and antibiotics by an 8-stranded b-barrel protein
To be Published
|
|
3V6A
 
 | |
3V6L
 
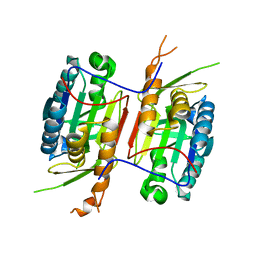 | | Crystal Structure of caspase-6 inactivation mutation | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
8FD0
 
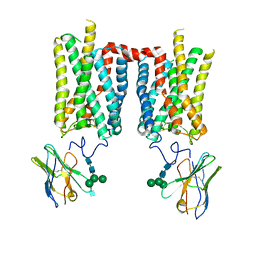 | | Crystal structure of bovine rod opsin in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb2, ... | | Authors: | Salom, D, Palczewski, K, Kiser, P.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain.
Nat Commun, 14, 2023
|
|
4FZM
 
 | | Crystal structure of the bacteriocin syringacin M from Pseudomonas syringae pv. tomato DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin, CALCIUM ION | | Authors: | Roszak, A.W, Grinter, R, Cogdell, J.R, Walker, D. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of the Lipid II-degrading Bacteriocin Syringacin M Suggests Unexpected Evolutionary Relationships between Colicin M-like Bacteriocins.
J.Biol.Chem., 287, 2012
|
|
4FZS
 
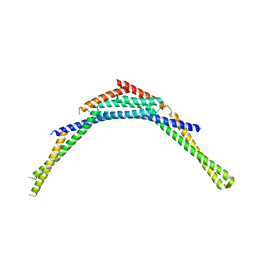 | | Structure of human SNX1 BAR domain | | Descriptor: | Sorting nexin-1 | | Authors: | van Weering, J.R.T, Sessions, R.B, Traer, C.J, Kloer, D.P, Bhatia, V.K, Stamou, D, Hurley, J.H, Cullen, P.J. | | Deposit date: | 2012-07-07 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insight into vesicle-to-tubule membrane remodeling by SNX-BAR proteins
To be Published
|
|
8FCZ
 
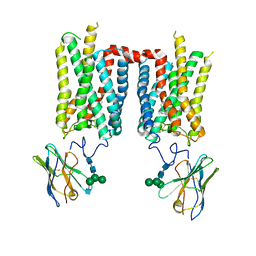 | | Crystal structure of ground-state rhodopsin in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb2, ... | | Authors: | Salom, D, Palczewski, K, Kiser, P.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain.
Nat Commun, 14, 2023
|
|
4G0C
 
 | | Neutron structure of acetazolamide-bound human carbonic anhydrase II reveal molecular details of drug binding. | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Fisher, S.Z, McKenna, R, Aggarwal, M. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Neutron diffraction of acetazolamide-bound human carbonic anhydrase II reveals atomic details of drug binding.
J.Am.Chem.Soc., 134, 2012
|
|
4FXB
 
 | | Crystal structure of CYP105N1 from Streptomyces coelicolor: a cytochrome P450 oxidase in the coelibactin siderophore biosynthetic pathway | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Hong, M.K, Lim, Y.R, Kim, J.K, Kim, D.H, Kang, L.W. | | Deposit date: | 2012-07-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of cytochrome P450 CYP105N1 from Streptomyces coelicolor, an oxidase in the coelibactin siderophore biosynthetic pathway
Arch.Biochem.Biophys., 528, 2012
|
|
4G0N
 
 | | Crystal Structure of wt H-Ras-GppNHp bound to the RBD of Raf Kinase | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Fetics, S.K, Kearney, B.M, Buhrman, G, Mattos, C. | | Deposit date: | 2012-07-09 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Allosteric Effects of the Oncogenic RasQ61L Mutant on Raf-RBD.
Structure, 23, 2015
|
|
4FYE
 
 | | Crystal structure of a Legionella phosphoinositide phosphatase, SidF | | Descriptor: | PHOSPHATE ION, SidF, inhibitor of growth family, ... | | Authors: | Hsu, F.S, Zhu, W, Brennan, L, Tao, L, Luo, Z.Q, Mao, Y. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G1P
 
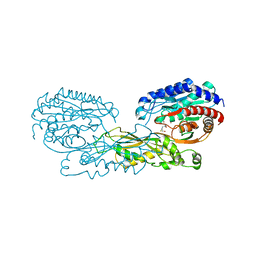 | | Structural and Mechanistic Basis of Substrate Recognition by Novel Di-peptidase Dug1p From Saccharomyces cerevisiae | | Descriptor: | CYSTEINE, Cys-Gly metallodipeptidase DUG1, GLYCINE, ... | | Authors: | Singh, A.K, Singh, M, Pandya, V.K, Singh, V, Mittal, M, Kumaran, S. | | Deposit date: | 2012-07-11 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Structural and Mechanistic Basis of Substrate Recognition by Novel Di-peptidase Dug1p From Saccromyces cerevesiae
To be published
|
|
3UYS
 
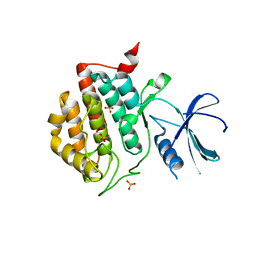 | | Crystal structure of apo human ck1d | | Descriptor: | Casein kinase I isoform delta, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2011-12-06 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor.
J.Med.Chem., 55, 2012
|
|
4G21
 
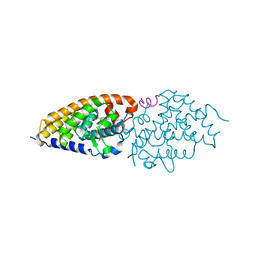 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
3UR3
 
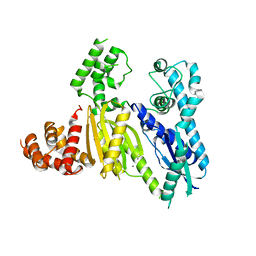 | | Structure of the Cmr2 subunit of the CRISPR RNA silencing complex | | Descriptor: | CALCIUM ION, Cmr2dHD, ZINC ION | | Authors: | Cocozaki, A.I, Ramia, N.F, Shao, Y, Hale, C.R, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of the Cmr2 Subunit of the CRISPR-Cas RNA Silencing Complex.
Structure, 20, 2012
|
|
4G2V
 
 | | Structure complex of LGN binding with FRMPD1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, G-protein-signaling modulator 2, ... | | Authors: | Shang, Y, Pan, Z, Wen, W, Wang, W, Zhang, M. | | Deposit date: | 2012-07-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of the interaction between LGN and Frmpd1
J.Mol.Biol., 425, 2013
|
|
3US3
 
 | | Recombinant rabbit skeletal calsequestrin-MPD complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Nissen, M.S, Munske, G.R, Kang, C. | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Glycosylation of skeletal calsequestrin: implications for its function.
J.Biol.Chem., 287, 2012
|
|
6MPR
 
 | |
4G1J
 
 | | Sortase C1 of GBS Pilus Island 1 | | Descriptor: | Sortase family protein | | Authors: | Cozzi, R, Prigozhin, D.M, Alber, T. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for group B streptococcus pilus 1 sortases C regulation and specificity.
Plos One, 7, 2012
|
|
4G1T
 
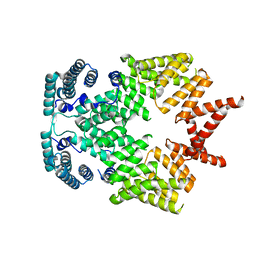 | | Crystal structure of interferon-stimulated gene 54 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 2 | | Authors: | Yang, Z, Liang, H, Zhou, Q, Li, Y, Chen, H, Ye, W, Chen, D, Fleming, J, Shu, H, Liu, Y. | | Deposit date: | 2012-07-11 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms.
Cell Res., 22, 2012
|
|
3V4G
 
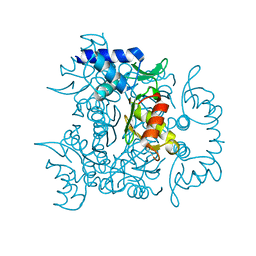 | | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6 | | Descriptor: | Arginine repressor | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E, Shuvalova, L, Winsor, J, Dubrovska, I, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6
To be Published
|
|
4G2F
 
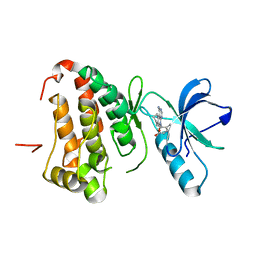 | | Human EphA3 kinase domain in complex with compound 7 | | Descriptor: | 1-amino-5-(5-hydroxy-2-methylphenyl)-7,8,9,10-tetrahydropyrimido[4,5-c]isoquinolin-6(5H)-one, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Discovery of a novel chemotype of tyrosine kinase inhibitors by fragment-based docking and molecular dynamics.
ACS MED.CHEM.LETT., 3, 2012
|
|
3V69
 
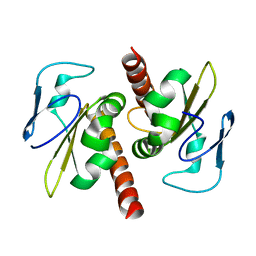 | | Filia-N crystal structure | | Descriptor: | Protein Filia | | Authors: | Wang, J, Xu, M, Zhu, K, Li, L, Liu, X. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-terminus of FILIA Forms an Atypical KH Domain with a Unique Extension Involved in Interaction with RNA.
Plos One, 7, 2012
|
|
