1VH7
 
 | |
1VHF
 
 | |
1VHZ
 
 | | Crystal structure of ADP compounds hydrolase | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP compounds hydrolase nudE | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIS
 
 | | Crystal structure of mevalonate kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate kinase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIZ
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | PcrB protein homolog, SODIUM ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHA
 
 | |
1VHU
 
 | | Crystal structure of a putative phosphoesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypothetical protein AF1521 | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VI5
 
 | | Crystal structure of ribosomal protein S2P | | Descriptor: | 30S ribosomal protein S2P | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHG
 
 | | Crystal structure of ADP compounds hydrolase | | Descriptor: | ADP compounds hydrolase nudE | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHT
 
 | |
1VI9
 
 | | Crystal structure of pyridoxamine kinase | | Descriptor: | BETA-MERCAPTOETHANOL, Pyridoxamine kinase, SULFATE ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VH6
 
 | | Crystal structure of a flagellar protein | | Descriptor: | Flagellar protein fliS | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHK
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yqeU | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VH2
 
 | |
1VH9
 
 | | Crystal structure of a putative thioesterase | | Descriptor: | Hypothetical protein ybdB | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
2I3H
 
 | | Structure of an ML-IAP/XIAP chimera bound to a 4-mer peptide (AVPW) | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AVPW peptide, ... | | Authors: | Fairbrother, W.J, Franklin, M.C. | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs.
Acs Chem.Biol., 1, 2006
|
|
2I3I
 
 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | (3R,6R,9AR)-2,2-DIMETHYL-6-[(N-METHYL-L-ALANYL)AMINO]-N-(3-METHYL-1-PHENYL-1H-PYRAZOL-5-YL)-5-OXO-2,3,5,6,9,9A-HEXAHYDRO[1,3]THIAZOLO[3,2-A]AZEPINE-3-CARBOXAMIDE, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fairbrother, W.J, Franklin, M.C. | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs.
Acs Chem.Biol., 1, 2006
|
|
2L4K
 
 | | Water refined solution structure of the human Grb7-SH2 domain in complex with the 10 amino acid peptide pY1139 | | Descriptor: | Growth factor receptor-bound protein 7, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Pias, S.C, Ivancic, M, Brescia, P.J, Johnson, D.L, Smith, D.E, Daly, R.J, Lyons, B.A. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-17 | | Last modified: | 2012-10-03 | | Method: | SOLUTION NMR | | Cite: | Water-Refined Solution Structure of the Human Grb7-SH2 Domain in Complex with the erbB2 Receptor Peptide pY1139.
Protein Pept.Lett., 19, 2012
|
|
2J2I
 
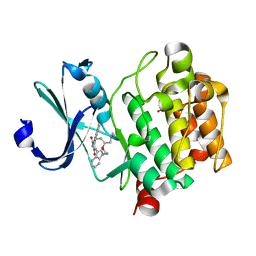 | | Crystal Structure of the humab PIM1 in complex with LY333531 | | Descriptor: | (9R)-9-[(DIMETHYLAMINO)METHYL]-6,7,10,11-TETRAHYDRO-9H,18H-5,21:12,17-DIMETHENODIBENZO[E,K]PYRROLO[3,4-H][1,4,13]OXADIA ZACYCLOHEXADECINE-18,20-DIONE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1, SULFATE ION | | Authors: | Debreczeni, J.E, Bullock, A.N, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, Knapp, S. | | Deposit date: | 2006-08-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
2IE2
 
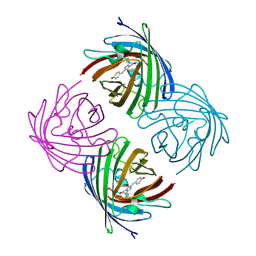 | |
2I7B
 
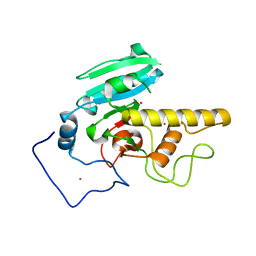 | |
2IBT
 
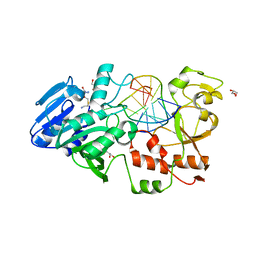 | | Crystal structure of the adenine-specific DNA methyltransferase M.TaqI complexed with the cofactor analog AETA and a 10 bp DNA containing 2-aminopurine at the target position and an abasic site analog at the target base partner position | | Descriptor: | 5'-D(*GP*AP*CP*AP*(3DR)P*CP*GP*(6MA)P*AP*C)-3', 5'-D(*GP*TP*TP*CP*GP*(2PR)P*TP*GP*TP*C)-3', 5'-DEOXY-5'-[2-(AMINO)ETHYLTHIO]ADENOSINE, ... | | Authors: | Lenz, T, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2006-09-12 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Aminopurine Flipped into the Active Site of the Adenine-Specific DNA Methyltransferase M.TaqI: Crystal Structures and Time-Resolved Fluorescence
J.Am.Chem.Soc., 129, 2007
|
|
3CIC
 
 | | Structure of BACE Bound to SCH709583 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7CIC
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 2-azanyl-N-[3-(trifluoromethyloxy)phenyl]ethanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
5CIC
 
 | |
