5CW1
 
 | | Proteinase K complexed with 4-iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, IODIDE ION, Proteinase K, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-12-30 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Iucrj, 3, 2016
|
|
7K6O
 
 | | Crystal structure of PI3Kalpha inhibitor 10-5429 | | Descriptor: | (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-N-methylpyrrolidine-1-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
1CVY
 
 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYPYPYBETADP)2 BOUND TO CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
4WYB
 
 | | Structure of the Bud6 flank domain in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Eck, M.J, Park, E, Zheng, W. | | Deposit date: | 2014-11-17 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Structure of a Bud6/Actin Complex Reveals a Novel WH2-like Actin Monomer Recruitment Motif.
Structure, 23, 2015
|
|
1XCK
 
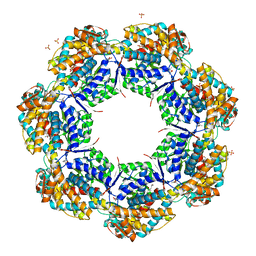 | | Crystal structure of apo GroEL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 60 kDa chaperonin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartolucci, C, Lamba, D, Grazulis, S, Manakova, E, Heumann, H. | | Deposit date: | 2004-09-02 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of wild-type chaperonin GroEL
J.Mol.Biol., 354, 2005
|
|
1FHX
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
8T8B
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site formyl-MAI-tripeptidyl-tRNA analog ACCA-IAMf at 2.65A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Thaler, J, Syroegin, E.A, Breuker, K, Polikanov, Y.S, Micura, R. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Practical Synthesis of N -Formylmethionylated Peptidyl-tRNA Mimics.
Acs Chem.Biol., 18, 2023
|
|
2Q44
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At1g77540 | | Descriptor: | BROMIDE ION, Uncharacterized protein At1g77540 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2GVI
 
 | |
8T8C
 
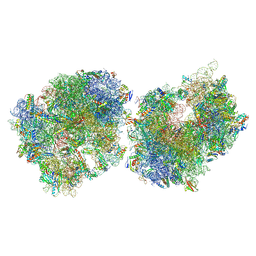 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site formyl-MFI-tripeptidyl-tRNA analog ACCA-IFMf at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Thaler, J, Syroegin, E.A, Breuker, K, Polikanov, Y.S, Micura, R. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Practical Synthesis of N -Formylmethionylated Peptidyl-tRNA Mimics.
Acs Chem.Biol., 18, 2023
|
|
6VGF
 
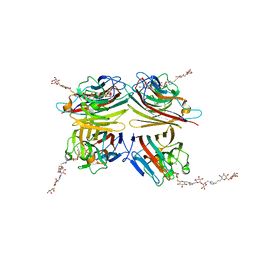 | | Peanut lectin complexed with divalent S-beta-D-thiogalactopyranosyl beta-D-glucopyranoside derivative (diSTGD) | | Descriptor: | (2S,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6R)-3,4,5-trihydroxy-6-{[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-({4-[({[(2S,3S,4S,5R,6S)-3,4,5-trihydroxy-6-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-2-yl]methyl}sulfanyl)methyl]-1H-1,2,3-triazol-1-yl}methyl)tetrahydro-2H-pyran-2-yl]oxy}tetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5H2I
 
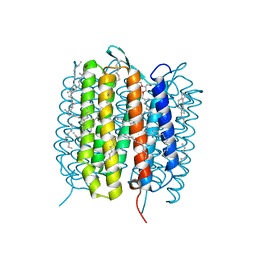 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 110 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5T96
 
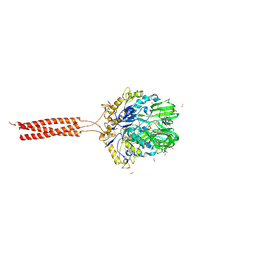 | | Crystal structure of the infectious salmon anemia virus (ISAV) HE viral receptor complex | | Descriptor: | 4-O-acetyl-5-acetamido-3,5-dideoxy-L-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Cook, J.D, Sultana, A, Lee, J.E. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the infectious salmon anemia virus receptor complex illustrates a unique binding strategy for attachment.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6YVE
 
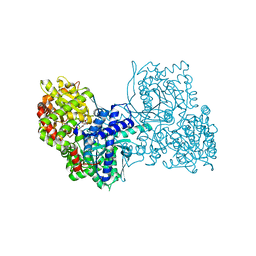 | | Glycogen phosphorylase b in complex with pelargonidin 3-O-beta-D-glucoside | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Drakou, C.E, Gardeli, C, Tsialtas, I, Alexopoulos, S, Mallouchos, A, Koulas, S, Tsagkarakou, A, Asimakopoulos, D, Leonidas, D.D, Psarra, A.M, Skamnaki, V.T. | | Deposit date: | 2020-04-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity Crystallography Reveals Binding of Pomegranate Juice Anthocyanins at the Inhibitor Site of Glycogen Phosphorylase: The Contribution of a Sugar Moiety to Potency and Its Implications to the Binding Mode.
J.Agric.Food Chem., 68, 2020
|
|
6YXX
 
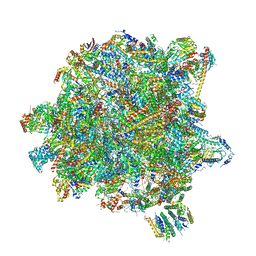 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
4TRF
 
 | |
1FE8
 
 | | CRYSTAL STRUCTURE OF THE VON WILLEBRAND FACTOR A3 DOMAIN IN COMPLEX WITH A FAB FRAGMENT OF IGG RU5 THAT INHIBITS COLLAGEN BINDING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, IMMUNOGLOBULIN IGG RU5, ... | | Authors: | Bouma, B, Huizinga, E.G, Schiphorst, M.E, Sixma, J.J, Kroon, J, Gros, P. | | Deposit date: | 2000-07-21 | | Release date: | 2001-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of the collagen-binding site of the von Willebrand factor A3-domain.
J.Biol.Chem., 276, 2001
|
|
2VX0
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N'-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-N-(3-MORPHOLIN-4-YLPHENYL)PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1FHW
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol(1,3,4,5,6)pentakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
7K71
 
 | | Crystal structure of PI3Kalpha inhibitor 4-0686 | | Descriptor: | 2-(morpholin-4-yl)[4,5'-bipyrimidin]-2'-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
1FN4
 
 | | CRYSTAL STRUCTURE OF FAB198, AN EFFICIENT PROTECTOR OF ACETYLCHOLINE RECEPTOR AGAINST MYASTHENOGENIC ANTIBODIES | | Descriptor: | MONOCLONAL ANTIBODY AGAINST ACETYLCHOLINE RECEPTOR | | Authors: | Poulas, K, Eliopoulos, E, Vatzaki, E, Navaza, J, Kontou, M. | | Deposit date: | 2000-08-21 | | Release date: | 2001-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Fab198, an efficient protector of the acetylcholine receptor against myasthenogenic antibodies.
Eur.J.Biochem., 268, 2001
|
|
3JCF
 
 | |
4A0K
 
 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | Descriptor: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5.93 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
6YXI
 
 | | Structure of Notum in complex with a 1-(3-Chlorophenyl)-2,5-dimethyl-1H-pyrrole-3-carboxylic acid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-chlorophenyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchia, L, Jones, E.Y, Ruza, R.R, Hillier, J, Zhao, Y. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
