3J41
 
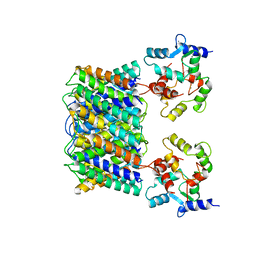 | | Pseudo-atomic model of the Aquaporin-0/Calmodulin complex derived from electron microscopy | | Descriptor: | CALCIUM ION, Calmodulin, Lens fiber major intrinsic protein | | Authors: | Reichow, S.L, Clemens, D.M, Freites, J.A, Nemeth-Cahalan, K.L, Heyden, M, Tobias, D.J, Hall, J.E, Gonen, T. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Allosteric mechanism of water-channel gating by Ca(2+)-calmodulin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6ZNO
 
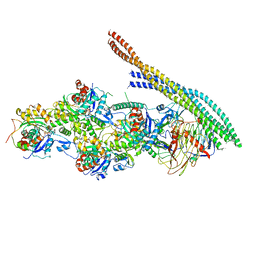 | | The pointed end complex of dynactin with the p150 projection docked | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Lacey, S.E, Carter, A.P. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
6ZNX
 
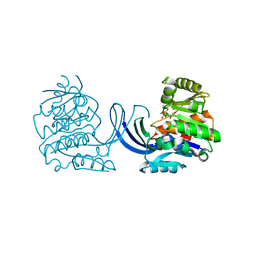 | | Ribokinase from Thermus Species | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribokinase | | Authors: | Timofeev, V.I, Abramchik, Y.A, Tuzova, E.S, Esipova, L.V, Mikheeva, O.O, Kostromina, M.A, Kuranova, I.P, Esipov, R.S. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ribokinase from Thermus Species
To Be Published
|
|
1XVP
 
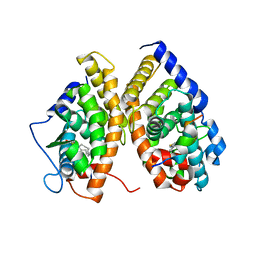 | | crystal structure of CAR/RXR heterodimer bound with SRC1 peptide, fatty acid and CITCO | | Descriptor: | 6-(4-CHLOROPHENYL)IMIDAZO[2,1-B][1,3]THIAZOLE-5-CARBALDEHYDE O-(3,4-DICHLOROBENZYL)OXIME, Orphan nuclear receptor NR1I3, PENTADECANOIC ACID, ... | | Authors: | Xu, R.X, Lambert, M.H, Wisely, B.B, Warren, E.N, Weinert, E.E, Waitt, G.M, Williams, J.D, Moore, L.B, Willson, T.M, Moore, J.T. | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Basis for Constitutive Activity in the Human CAR/RXRalpha Heterodimer.
Mol.Cell, 16, 2004
|
|
4YWL
 
 | |
6ZOK
 
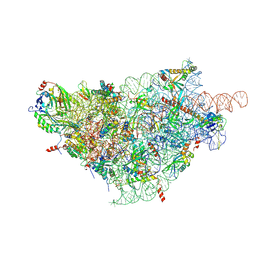 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZPK
 
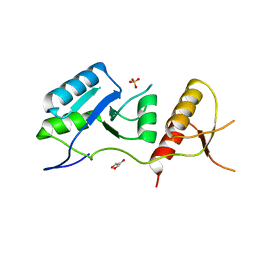 | | Crystal structure of the unconventional kinetochore protein Trypanosoma brucei KKT4 BRCT domain | | Descriptor: | GLYCEROL, SULFATE ION, Trypanosoma brucei KKT4 463-645 | | Authors: | Ludzia, P, Lowe, E.D, Marciano, G, Mohammed, S, Redfield, C, Akiyoshi, B. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.
Structure, 29, 2021
|
|
6BDX
 
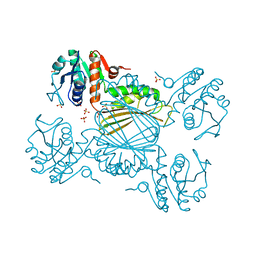 | | 4-hydroxy tetrahydrodipicolinate reductase from Neisseria gonorrhoeae | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, SULFATE ION | | Authors: | Pote, S.S, Pye, S.E, Sheahan, T.E, Chruszcz, M. | | Deposit date: | 2017-10-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 4-Hydroxy-tetrahydrodipicolinate reductase from Neisseria gonorrhoeae - structure and interactions with coenzymes and substrate analog.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6Z3R
 
 | | Structure of SMG1-8-9 kinase complex bound to UPF1-LSQ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Langer, L.M, Gat, Y, Conti, E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of substrate-bound SMG1-8-9 kinase complex reveals molecular basis for phosphorylation specificity.
Elife, 9, 2020
|
|
6ETB
 
 | | Aerobic S262Y mutation of E. coli FLRD core | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Romao, C.V, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | A tyrosine mutation in E. coli flavodiiron protein increases radiation sensitivity in the crystal structure
To Be Published
|
|
1KHQ
 
 | | ORTHORHOMBIC FORM OF PAPAIN/ZLFG-DAM COVALENT COMPLEX | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
4RED
 
 | | Crystal structure of human AMPK alpha1 KD-AID with K43A mutation | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
6ZDV
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with Chromone 5d | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Verdon, G, Jespers, W, Azuaje, J, Majellaro, M, Keranen, H, Garcia-mera, X, Congreve, M, Deflorian, F, de Graaf, C, Zhukov, A, Dore, A, Mason, J, Aqvist, J, Cooke, R, Sotelo, E, Gutierrez-de-Teran, H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | X-Ray Crystallography and Free Energy Calculations Reveal the Binding Mechanism of A 2A Adenosine Receptor Antagonists.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5H2J
 
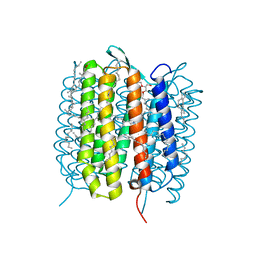 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 290 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
2CLP
 
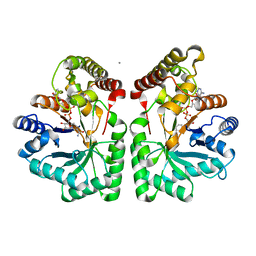 | | Crystal structure of human aflatoxin B1 aldehyde reductase member 3 | | Descriptor: | AFLATOXIN B1 ALDEHYDE REDUCTASE MEMBER 3, CALCIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Debreczeni, J.E, Marsden, B.D, Johansson, C, Kavanagh, K, Guo, K, Smee, C, Gileadi, O, Turnbull, A, Papagrigoriou, E, von Delft, F, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Oppermann, U. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Aflatoxin B1 Aldehyde Reductase Member 3
To be Published
|
|
6ZHK
 
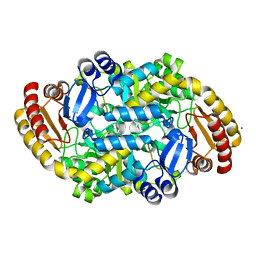 | | Crystal structure of adenosylmethionine-8-amino-7-oxononanoate aminotransferase from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, MAGNESIUM ION | | Authors: | Boyko, K.M, Nikolaeva, T.N, Stekhanova, T.N, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2020-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-Dimensional Structure of Thermostable D-Amino Acid Transaminase from the Archaeon Methanocaldococcus jannaschii DSM 2661
Crystallography Reports, 2021
|
|
5H2P
 
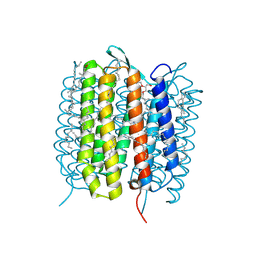 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 657 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
8F21
 
 | | Structure of a 30mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F0A
 
 | | Client-bound structure of a DegP trimer within a 12mer cage | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
5ACF
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
8F1T
 
 | | Structure of an 18mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (12.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F1U
 
 | | Structure of a 24mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (13.8 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
7PFS
 
 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide ((1R)-1-Amino-3-phenylpropyl){2-([1,1:3,1-terphenyl]-5-ylmethyl)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)-amino]-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
2CCH
 
 | | The crystal structure of CDK2 cyclin A in complex with a substrate peptide derived from CDC modified with a gamma-linked ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6 HOMOLOG, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of the Phospho-Cdk2/Cyclin a Recruitment Site in Substrate Recognition
J.Biol.Chem., 281, 2006
|
|
1VKC
 
 | | Putative acetyl transferase from Pyrococcus furiosus | | Descriptor: | IODIDE ION, putative acetyl transferase | | Authors: | Habel, J.E, Liu, Z.-J, Tempel, W, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Putative acetyl transferase from Pyrococcus furiosus
To be published
|
|
