6ZHJ
 
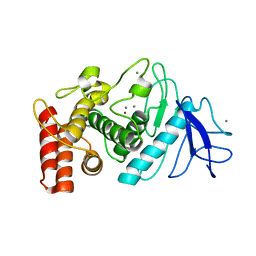 | | 3D electron diffraction structure of thermolysin from Bacillus thermoproteolyticus | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.26 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8SLZ
 
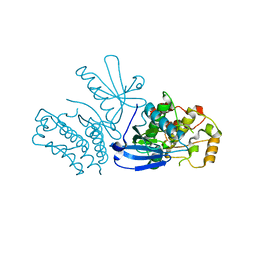 | |
8SM3
 
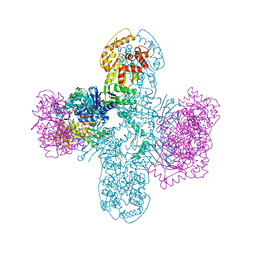 | | Structure of Bacillus cereus VD045 Gabija GajA-GajB Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB, SULFATE ION | | Authors: | Antine, S.P, Mooney, S.E, Johnson, A.G, Kranzusch, P.J. | | Deposit date: | 2023-04-25 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
6YSK
 
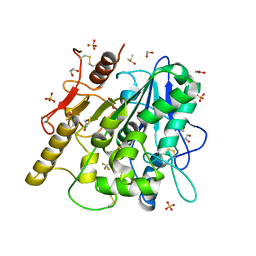 | | 1-phenylpyrroles and 1-enylpyrrolidines as inhibitors of Notum | | Descriptor: | (3~{S})-1-[4-chloranyl-3-(trifluoromethyl)phenyl]pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
5GM8
 
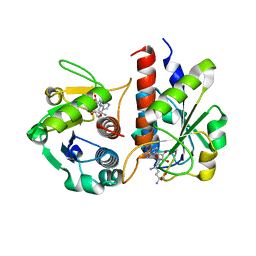 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | SINEFUNGIN, tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
5GMC
 
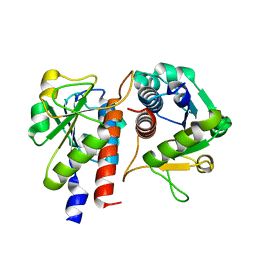 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
6ZK8
 
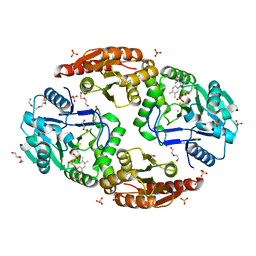 | | Native crystal structure of anaerobic F420H2-Oxidase from Methanothermococcus thermolithotrophicus at 1.8A resolution | | Descriptor: | Coenzyme F420H2 oxidase (FprA), DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Engilberge, S, Wagner, T, Carpentier, P, Girard, E, Shima, S. | | Deposit date: | 2020-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Krypton-derivatization highlights O 2 -channeling in a four-electron reducing oxidase.
Chem.Commun.(Camb.), 56, 2020
|
|
5H2M
 
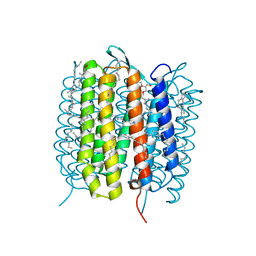 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 13.8 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
6E9Q
 
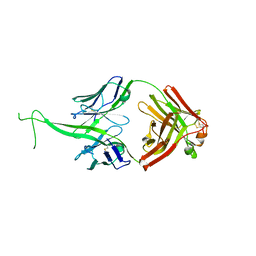 | |
1GID
 
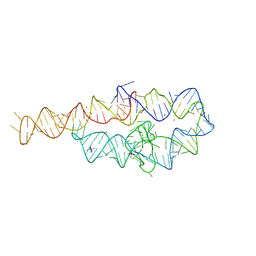 | | CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA PACKING | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, P4-P6 RNA RIBOZYME DOMAIN | | Authors: | Cate, J.H, Gooding, A.R, Podell, E, Zhou, K, Golden, B.L, Kundrot, C.E, Cech, T.R, Doudna, J.A. | | Deposit date: | 1996-08-22 | | Release date: | 1996-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a group I ribozyme domain: principles of RNA packing.
Science, 273, 1996
|
|
1GLM
 
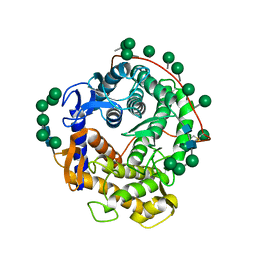 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | Descriptor: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-04-25 | | Release date: | 1994-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
1GM7
 
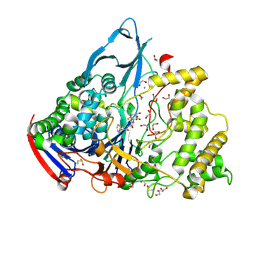 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
4Y6G
 
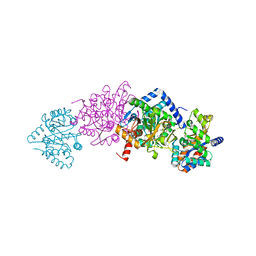 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor in the alpha-site and beta-site. | | Descriptor: | 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2015-02-13 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
2MSF
 
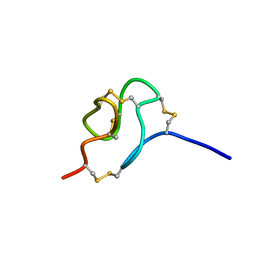 | | NMR SOLUTION STRUCTURE OF SCORPION VENOM TOXIN Ts11 (TsPep1) FROM Tityus serrulatus | | Descriptor: | Peptide TsPep1 | | Authors: | Maiti, M, Lescrinier, E, Herdewijn, P, Cremonez, C.M, Peigneur, S, Cassoli, J.S, Dutra, A.A.A, Waelkens, E, Pimenta, A.M.C, De Lima, M.H, Tytgat, J, Arantes, E.C. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Elucidation of Peptide Ts11 Shows Evidence of a Novel Subfamily of Scorpion Venom Toxins.
Toxins (Basel), 8, 2016
|
|
6UH3
 
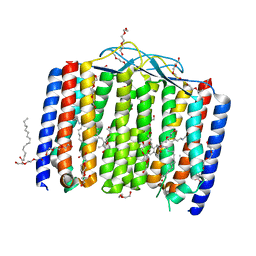 | | Crystal structure of bacterial heliorhodopsin 48C12 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heliorhodopsin, PALMITIC ACID, ... | | Authors: | Lu, Y, Zhou, X.E, Gao, X, Xia, R, Xu, Z, Wang, N, Leng, Y, Melcher, K, Xu, H.E, He, Y. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of heliorhodopsin 48C12.
Cell Res., 30, 2020
|
|
6ARK
 
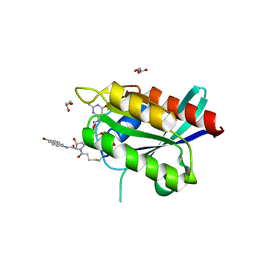 | | Crystal Structure of compound 10 covalently bound to K-Ras G12C | | Descriptor: | (3R)-N-(6-bromonaphthalen-2-yl)-3-hydroxy-1-propanoyl-L-prolinamide, GLYCEROL, GTPase KRas, ... | | Authors: | Nnadi, C.I, Jenkins, M.L, Gentile, D.R, Bateman, L.A, Zaidman, D, Balius, T.E, Nomura, D.K, Burke, J.E, Shokat, K.M, London, N. | | Deposit date: | 2017-08-22 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel K-Ras G12C Switch-II Covalent Binders Destabilize Ras and Accelerate Nucleotide Exchange.
J Chem Inf Model, 58, 2018
|
|
1GMJ
 
 | | The structure of bovine IF1, the regulatory subunit of mitochondrial F-ATPase | | Descriptor: | ATPASE INHIBITOR | | Authors: | Cabezon, E, Runswick, M.J, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2001-09-14 | | Release date: | 2002-01-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Bovine If(1), the Regulatory Subunit of Mitochondrial F-ATPase.
Embo J., 20, 2001
|
|
4Y7O
 
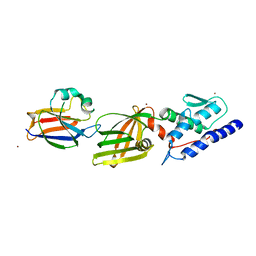 | | T6SS protein TssM C-terminal domain (869-1107) from EAEC | | Descriptor: | Type VI secretion protein IcmF, Type VI secretion system protein VasD, ZINC ION | | Authors: | Nguyen, V.S, Spinelli, S, Durand, E, Roussel, A, Cambillau, C. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Biogenesis and structure of a type VI secretion membrane core complex.
Nature, 523, 2015
|
|
5F97
 
 | | Blood group antigen binding adhesin BabA of Helicobacter pylori strain A730 in complex with blood group A type 1 hexasaccharide | | Descriptor: | Adhesin binding fucosylated histo-blood group antigen,Adhesin,Adhesin binding fucosylated histo-blood group antigen, Nanbody Nb-ER19, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Moonens, K, Gideonsson, P, Subedi, S, Romao, E, Oscarson, S, Muyldermans, S, Boren, T, Remaut, H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Cell Host Microbe, 19, 2016
|
|
6Z43
 
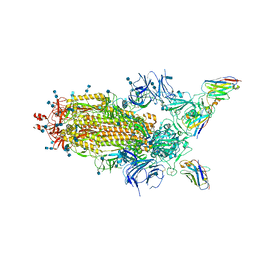 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
1LLS
 
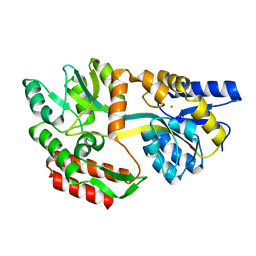 | | CRYSTAL STRUCTURE OF UNLIGANDED MALTOSE BINDING PROTEIN WITH XENON | | Descriptor: | Maltose-binding periplasmic protein, XENON | | Authors: | Rubin, S.M, Lee, S.-Y, Ruiz, E.J, Pines, A, Wemmer, D.E. | | Deposit date: | 2002-04-30 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DETECTION AND CHARACTERIZATION OF XENON-BINDING SITES IN PROTEINS BY 129XE NMR SPECTROSCOPY
J.MOL.BIOL., 322, 2002
|
|
6EKR
 
 | |
4Y8L
 
 | | Yeast 20S proteasome in complex with Ac-APLL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-APLL-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
5C0W
 
 | |
6Z2O
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, O-glycan protease, ZINC ION | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
