5AKV
 
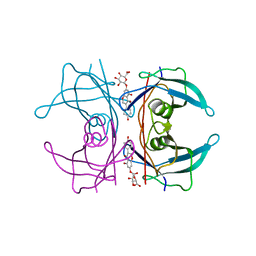 | | Transthyretin binding heterogeneity and anti-amyloidogenic activity of natural polyphenols and their metabolites: genistein-7-O- glucuronide | | Descriptor: | Genistein-7-O-glucuronide, TRANSTHYRETIN | | Authors: | Florio, P, Foll, C, Cianci, M, Del Rio, D, Zanotti, G, Berni, R. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Transthyretin Binding Heterogeneity and Anti-Amyloidogenic Activity of Natural Polyphenols and Their Metabolites
J.Biol.Chem., 290, 2015
|
|
6ZUS
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular protein 11-1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
6ZUQ
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | Extracellular protein 11-1, GLYCEROL, ZINC ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7AD5
 
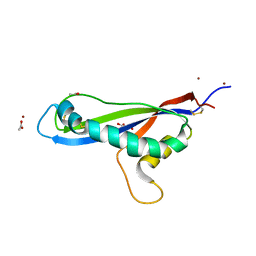 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | ACETATE ION, Avirulence protein LmJ1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
3DJS
 
 | |
3DJZ
 
 | |
4P5K
 
 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between Allergy and Autoimmunity | | Descriptor: | HLA class II histocompatibility antigen, DP alpha 1 chain, RAS peptide,HLA class II histocompatibility antigen, ... | | Authors: | Clayton, G.M, Wang, Y, Crawford, F, Novikov, A, Wimberly, B.T, Kieft, J.S, Falta, M.T, Bowerman, N.A, Marrack, P, Fontenot, A.P, Dai, S, Kappler, J.W. | | Deposit date: | 2014-03-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
7SZZ
 
 | | Structure of the smaller diameter PSMalpha3 nanotubes | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Beltran, L.C, Kreutzberger, M.A, Wang, S, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-11-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T8U
 
 | |
7T0X
 
 | | Structure of the larger diameter PSMalpha3 nanotube | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Kreutzberger, M.A, Wang, S, Beltran, L.C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-11-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6PX7
 
 | |
2RGK
 
 | | Functional annotation of Escherichia coli yihS-encoded protein | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Uncharacterized sugar isomerase yihS | | Authors: | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-03 | | Release date: | 2008-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
6PX8
 
 | |
4B9B
 
 | | The structure of the omega aminotransferase from Pseudomonas aeruginosa | | Descriptor: | BETA-ALANINE-PYRUVATE TRANSAMINASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sayer, C, Isupov, M.N, Westlake, A, Littlechild, J.A. | | Deposit date: | 2012-09-03 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6DQP
 
 | |
6DQI
 
 | |
6DQO
 
 | |
4BA5
 
 | | Crystal structure of omega-transaminase from Chromobacterium violaceum | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, AMINOTRANSFERASE, SULFATE ION | | Authors: | Sayer, C, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2012-09-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4B98
 
 | | The structure of the omega aminotransferase from Pseudomonas aeruginosa | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, BETA-ALANINE--PYRUVATE TRANSAMINASE, CALCIUM ION, ... | | Authors: | Sayer, C, Isupov, M.N, Westlake, A, Littlechild, J.A. | | Deposit date: | 2012-09-03 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8AZ8
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 2-(benzylamino)ethan-1-ol | | Descriptor: | 2-[(phenylmethyl)amino]ethanol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
4AO4
 
 | | Structural Determinants of the beta-Selectivity of a Bacterial Aminotransferase | | Descriptor: | (3R)-3-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]-5-METHYLHEXANOIC ACID, 1,2-ETHANEDIOL, Beta-transaminase | | Authors: | Wybenga, G.G, Crismaru, C.G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2012-03-23 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of the beta-selectivity of a bacterial aminotransferase.
J. Biol. Chem., 287, 2012
|
|
8ORS
 
 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
8ORH
 
 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
4C6Y
 
 | | Ancestral PNCA (last common ancestors of Gram-positive and Gram- negative bacteria) beta-lactamase class A | | Descriptor: | ACETATE ION, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Phenotypic Comparisons of Consensus Variants Versus Laboratory Resurrections of Precambrian Proteins.
Proteins, 82, 2014
|
|
