5XGS
 
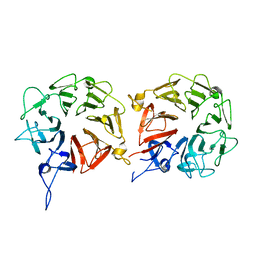 | | Crystal structure of human WBSCR16 | | Descriptor: | RCC1-like G exchanging factor-like protein | | Authors: | Koyama, M, Sasaki, N, Matsuura, Y. | | Deposit date: | 2017-04-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human WBSCR16, an RCC1-like protein in mitochondria
Protein Sci., 26, 2017
|
|
5XVC
 
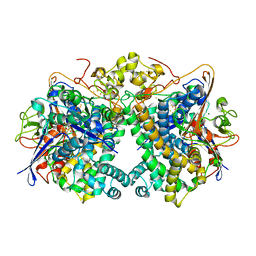 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XW4
 
 | |
5XW5
 
 | |
5XOJ
 
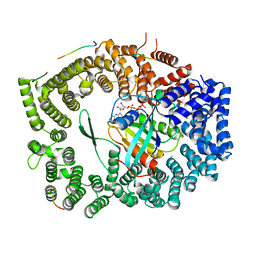 | | Crystal structure of Xpo1p-PKI-Nup42p-Gsp1p-GTP complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koyama, M, Shirai, N, Matsuura, Y. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Xpo1p nuclear export complex bound to the SxFG/PxFG repeats of the nucleoporin Nup42p
Genes Cells, 22, 2017
|
|
5X8N
 
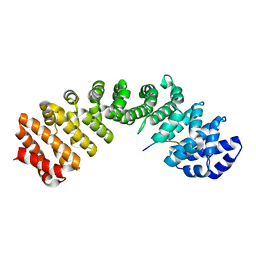 | |
5XVB
 
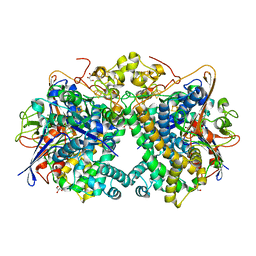 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | Descriptor: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XZX
 
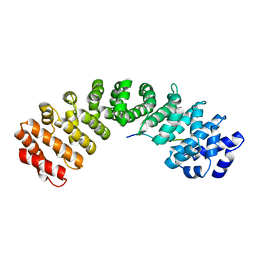 | |
3VOQ
 
 | |
2Z6R
 
 | | Crystal structure of Lys49 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N. | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
3VYC
 
 | |
3W3X
 
 | | Crystal structure of Kap121p bound to Pho4p | | Descriptor: | Importin subunit beta-3, Phosphate system positive regulatory protein PHO4 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3Y
 
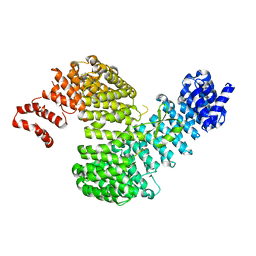 | | Crystal structure of Kap121p bound to Nup53p | | Descriptor: | Importin subunit beta-3, Nucleoporin NUP53 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3T
 
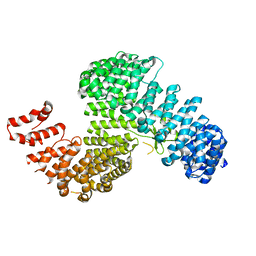 | | Crystal structure of Kap121p | | Descriptor: | Importin subunit beta-3 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3V
 
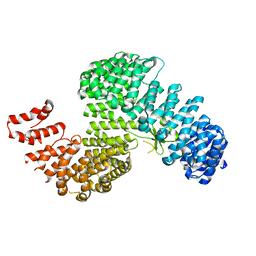 | |
3WUV
 
 | | Structure basis of inactivating cell abscission with chimera peptide 2 | | Descriptor: | Centrosomal protein of 55 kDa, peptide from Programmed cell death 6-interacting protein | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3W3U
 
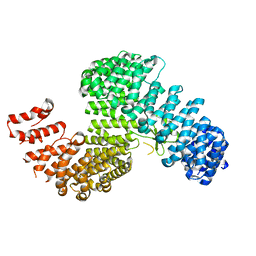 | |
3W3Z
 
 | | Crystal structure of Kap121p bound to RanGTP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin subunit beta-3, ... | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3X2Y
 
 | | Crystal structure of metallo-beta-lactamase H8A from Thermotoga maritima | | Descriptor: | NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of metallo-beta-lactamase H8A from Thermotoga maritima
To be Published
|
|
3X30
 
 | | Crystal structure of metallo-beta-lactamase from Thermotoga maritima | | Descriptor: | MANGANESE (II) ION, NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Crystal structure of metallo-beta-lactamase from Thermotoga maritima
To be Published
|
|
3WYF
 
 | | Crystal structure of Xpo1p-Yrb2p-Gsp1p-GTP complex | | Descriptor: | Exportin-1, GUANOSINE-5'-TRIPHOSPHATE, Gsp1p, ... | | Authors: | Koyama, M, Shirai, N, Matsuura, Y. | | Deposit date: | 2014-08-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural insights into how yrb2p accelerates the assembly of the xpo1p nuclear export complex
Cell Rep, 9, 2014
|
|
3X2X
 
 | | Crystal structure of metallo-beta-lactamase H48A from Thermotoga maritima | | Descriptor: | MANGANESE (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Crystal structure of metallo-beta-lactamase H48A from Thermotoga maritima
To be Published
|
|
3X3U
 
 | |
3X2Z
 
 | | Crystal structure of metallo-beta-lactamase in complex with nickel from Thermotoga maritima | | Descriptor: | NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of metallo-beta-lactamase in complex with nickel from Thermotoga maritima
To be Published
|
|
