8SPZ
 
 | | Crystal structure of Bax core domain BH3-groove dimer - hexameric fraction with dioctanoyl phosphatidylserine | | Descriptor: | Apoptosis regulator BAX, SULFATE ION | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E, Miller, M.S. | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SRX
 
 | | Crystal structure of BAK-BAX heterodimer with lysoPC | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, Bcl-2 homologous antagonist/killer, ... | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M, Miller, M.S. | | Deposit date: | 2023-05-07 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
5LAK
 
 | | Ligand-bound structure of Cavally Virus 3CL Protease | | Descriptor: | 3Cl Protease, BEZ-TYR-TYR-ASN-ECC Peptide inhibitor, PENTAETHYLENE GLYCOL, ... | | Authors: | Kanitz, M, Heine, A, Diederich, W.E. | | Deposit date: | 2016-06-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus.
Virology, 533, 2019
|
|
8SRY
 
 | | Crystal structure of BAK-BAX heterodimer with C12E8 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, ... | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M, Miller, M.S. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
5KNE
 
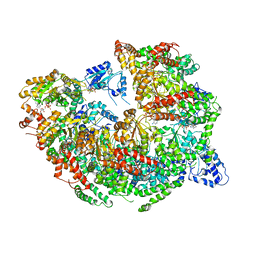 | | CryoEM Reconstruction of Hsp104 Hexamer | | Descriptor: | Heat shock protein 104, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yokom, A.L, Gates, S.N, Jackrel, M.E, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2016-06-28 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.64 Å) | | Cite: | Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5KRI
 
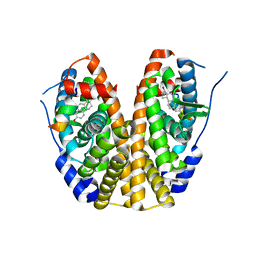 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 16b-benzyl 17b-estradiol | | Descriptor: | (8~{R},9~{S},13~{S},14~{S},16~{R},17~{S})-13-methyl-16-(phenylmethyl)-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthrene-3,17-diol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5AMN
 
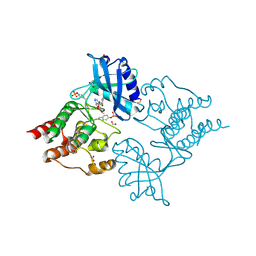 | | The Discovery of 2-Substituted Phenol Quinazolines as Potent and Selective RET Kinase Inhibitors | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Newton, R, Bowler, K, Burns, E.M, Chapman, P, Fairweather, E, Fritzl, S, Goldberg, K, Hamilton, N.M, Holt, S.V, Hopkins, G.V, Jones, S.D, Jordan, A.M, Lyons, A, McDonald, N.Q, Maguire, L.A, Mould, D.P, Purkiss, A.G, Small, H.F, Stowell, A, Thomson, G.J, Waddell, I.D, Waszkowycz, B, Watson, A.J, Ogilvie, D.J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The discovery of 2-substituted phenol quinazolines as potent RET kinase inhibitors with improved KDR selectivity.
Eur J Med Chem, 112, 2016
|
|
6EQD
 
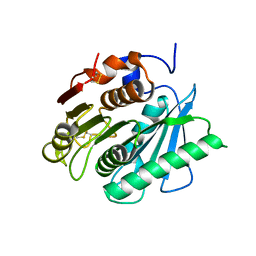 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis collected at long wavelength | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5L7F
 
 | | Crystal structure of MMP12 mutant K421A in complex with RXP470.1 conjugated with fluorophore Cy5,5 in space group P21. | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Tepshi, L, Bordenave, T, Rouanet-Mehouas, C, Devel, L, Dive, V, Stura, E.A. | | Deposit date: | 2016-06-03 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and in Vitro and in Vivo Evaluation of MMP-12 Selective Optical Probes.
Bioconjug.Chem., 27, 2016
|
|
4XIW
 
 | | Carbonic anhydrase Cah3 from Chlamydomonas reinhardtii in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, alpha type, ... | | Authors: | Hainzl, T, Grundstrom, C, Benlloch, R, Shevela, D, Shutova, T, Messinger, J, Samuelsson, G, Sauer-Eriksson, A.E. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Functional Characterization of Photosystem II-Associated Carbonic Anhydrase CAH3 in Chlamydomonas reinhardtii.
Plant Physiol., 167, 2015
|
|
6EIG
 
 | | Crystal structure of N24Q/C128T mutant of Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, EICOSANE, ... | | Authors: | Kovalev, K, Borshchevskiy, V, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
5KOH
 
 | | Nitrogenase MoFeP from Gluconacetobacter diazotrophicus in dithionite reduced state | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Owens, C.P, Tezcan, F.A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Tyrosine-Coordinated P-Cluster in G. diazotrophicus Nitrogenase: Evidence for the Importance of O-Based Ligands in Conformationally Gated Electron Transfer.
J.Am.Chem.Soc., 138, 2016
|
|
5KU1
 
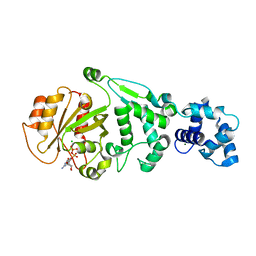 | | hMiro1 EF hand and cGTPase domains in the GDP-bound state | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
7ZBE
 
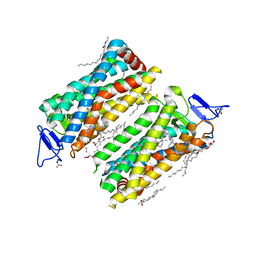 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SwissFEL) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
3L3X
 
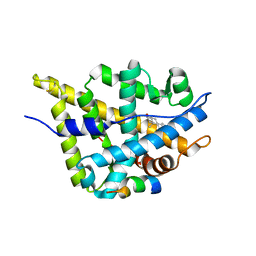 | | Crystal structure of DHT-bound androgen receptor in complex with the first motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
5DKR
 
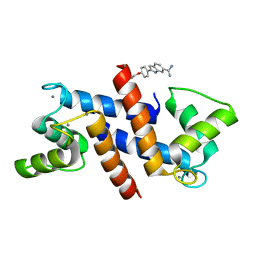 | | Crystal Structure of Calcium-loaded S100B bound to SBi29 | | Descriptor: | 2-[4-(4-carbamimidoylphenoxy)phenyl]-1H-indole-6-carboximidamide, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
8FDQ
 
 | |
6FQB
 
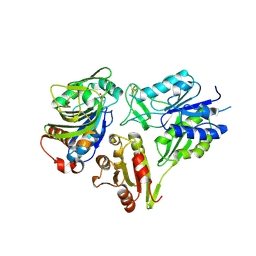 | | MurT/GatD peptidoglycan amidotransferase complex from Streptococcus pneumoniae R6 | | Descriptor: | Cobyric acid synthase, GLUTAMINE, Mur ligase family protein | | Authors: | Morlot, C, Contreras-Martel, C, Leisico, F, Straume, D, Peters, K, Hegnar, O.A, Simon, N, Villard, A.M, Breukink, E, Gravier-Pelletier, C, Le Corre, L, Vollmer, W, Pietrancosta, N, Havarstein, L.S, Zapun, A. | | Deposit date: | 2018-02-13 | | Release date: | 2018-08-22 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
2VBA
 
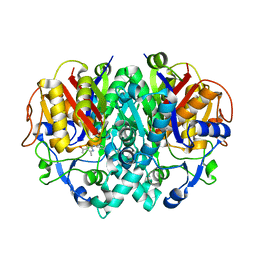 | | beta-ketoacyl-ACP synthase I (KAS) from E. coli with bound amino- thiazole inhibitor | | Descriptor: | 2-PHENYLAMINO-4-METHYL-5-ACETYL THIAZOLE, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1 | | Authors: | Pappenberger, G, Schulz-Gasch, T, Bailly, J, Hennig, M. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-Assisted Discovery of an Aminothiazole Derivative as a Lead Molecule for Inhibition of Bacterial Fatty-Acid Synthesis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2VFC
 
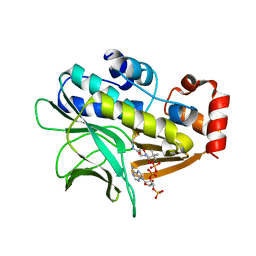 | | The structure of Mycobacterium marinum arylamine N-acetyltransferase in complex with CoA | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE, COENZYME A | | Authors: | Fullam, E, Westwood, I.M, Anderton, M.C, Lowe, E.D, Sim, E, Noble, M.E.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Divergence of Cofactor Recognition Across Evolution: Coenzyme a Binding in a Prokaryotic Arylamine N-Acetyltransferase.
J.Mol.Biol., 375, 2008
|
|
6G9B
 
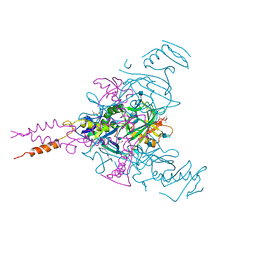 | | Crystal structure of Ebolavirus glycoprotein in complex with imipramine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, ... | | Authors: | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
5L79
 
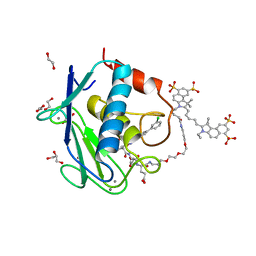 | | Crystal structure of MMP12 in complex with RXP470.1 conjugated with fluorophore Cy5,5 in space group P21212. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CY5.5-PEG2, ... | | Authors: | Tepshi, L, Bordenave, T, Devel, L, Dive, V, Stura, E.A. | | Deposit date: | 2016-06-02 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis and in Vitro and in Vivo Evaluation of MMP-12 Selective Optical Probes.
Bioconjug.Chem., 27, 2016
|
|
3KSA
 
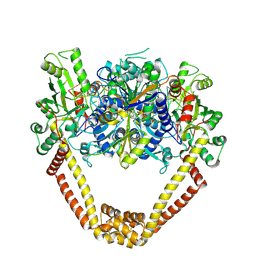 | | Detailed structural insight into the DNA cleavage complex of type IIA topoisomerases (cleaved form) | | Descriptor: | 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3', 5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2009-11-21 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
6GAL
 
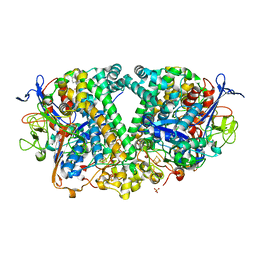 | | Structure of fully reduced Hydrogenase (Hyd-1) variant E28Q collected at pH 10 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6TMN
 
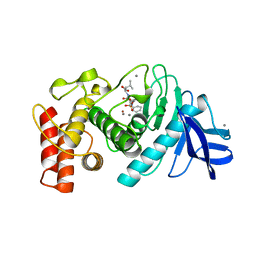 | | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond | | Descriptor: | CALCIUM ION, N-[(2R,4S)-4-hydroxy-2-(2-methylpropyl)-4-oxido-7-oxo-9-phenyl-3,8-dioxa-6-aza-4-phosphanonan-1-oyl]-L-leucine, THERMOLYSIN, ... | | Authors: | Tronrud, D.E, Holden, H.M, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond.
Science, 235, 1987
|
|
