4V4X
 
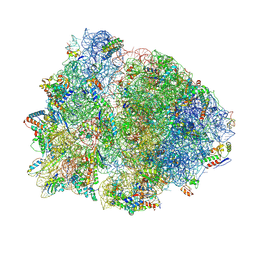 | | Crystal structure of the 70S Thermus thermophilus ribosome showing how the 16S 3'-end mimicks mRNA E and P codons. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
4FSP
 
 | |
4FT6
 
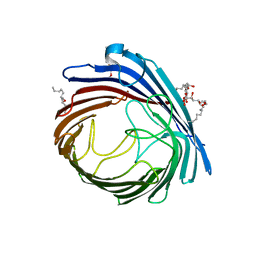 | |
5HVD
 
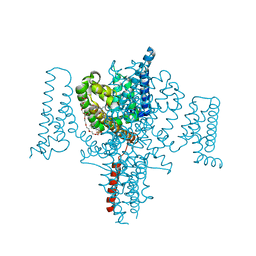 | | Full length Open-form Sodium Channel NavMs I218C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Booker, J.E, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2016-01-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The complete structure of an activated open sodium channel.
Nat Commun, 8, 2017
|
|
5HXF
 
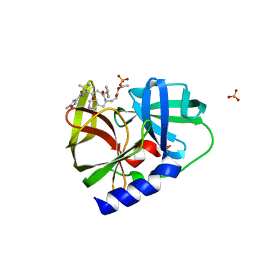 | |
4G3S
 
 | |
4WAL
 
 | | Crystal structure of selenomethionine Msl5 protein in complex with RNA at 2.2 A | | Descriptor: | Branchpoint-bridging protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
4GEK
 
 | | Crystal Structure of wild-type CmoA from E.coli | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, SULFATE ION, tRNA (cmo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bonanno, J.B, Bhosle, R, Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Nature, 498, 2013
|
|
5I6Y
 
 | |
5I89
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02857790 | | Descriptor: | (4R)-6-(3-cyclopropyl-1-methyl-1H-indazol-5-yl)-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Setser, J.W, Poy, F, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5IPG
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure FFT-butyl (Hyperoxodized by t-butyl hydroperoxide) | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
4GEI
 
 | | N-terminal domain of VDUP-1 | | Descriptor: | Thioredoxin-interacting protein | | Authors: | Polekhina, G, Kok, S.F, Ascher, D.B, Waltham, M. | | Deposit date: | 2012-08-02 | | Release date: | 2013-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the N-terminal domain of human thioredoxin-interacting protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5I83
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02773986 | | Descriptor: | (4R)-4-methyl-7-[(1R)-1-phenylethoxy]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, CREB-binding protein, THIOCYANATE ION | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
4UW5
 
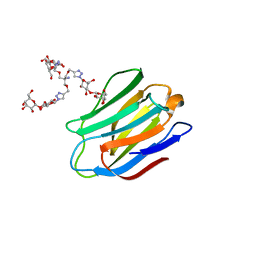 | | Human galectin-7 in complex with a galactose based dendron D2-2. | | Descriptor: | DENDRON D2-1, HUMAN GALECTIN-7 | | Authors: | Ramaswamy, S, Sleiman, M.H, Masuyer, G, Arbez-Gindre, C, Micha-Screttas, M, Calogeropoulou, T, Steele, B.R, Acharya, K.R. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis of Multivalent Galactose-Based Dendrimer Recognition by Human Galectin-7.
FEBS J., 282, 2015
|
|
4GEJ
 
 | | N-terminal domain of VDUP-1 | | Descriptor: | CALCIUM ION, Thioredoxin-interacting protein | | Authors: | Polekhina, G, Kok, S.F, Ascher, D.B, Waltham, M. | | Deposit date: | 2012-08-02 | | Release date: | 2013-02-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the N-terminal domain of human thioredoxin-interacting protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5IMD
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F4 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, OXYGEN ATOM, ... | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-06 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5INY
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F8 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
4FUO
 
 | |
4UW3
 
 | | Human galectin-7 in complex with a galactose based dendron D1. | | Descriptor: | DENDRON-D1, HUMAN GALECTIN-7 | | Authors: | Ramaswamy, S, Sleiman, M.H, Masuyer, G, Arbez-Gindre, C, Micha-Screttas, M, Calogeropoulou, T, Steele, B.R, Acharya, K.R. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structural Basis of Multivalent Galactose-Based Dendrimer Recognition by Human Galectin-7.
FEBS J., 282, 2015
|
|
5I9P
 
 | |
4GF7
 
 | | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Sawa, Y, Yagi, T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
5IIU
 
 | | Crystal structure of Equine Serum Albumin in the presence of 10 mM zinc at pH 6.9 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Cymborowski, M.T, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
4URP
 
 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | Descriptor: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | Authors: | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
4GH0
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5IOX
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure LUss | | Descriptor: | Bacterioferritin comigratory protein | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
